General strategy for analysis:
Define formula via scientific questions + confounders.
Define type of GLM (lm, logistic, Poisson).
Blocks in data -> Random effects, start with random intercept.
Fit this base model, then do residual checks for
Wrong functional form -> Change fitted function.
Wrong distribution-> Transformation or GLM adjustment.
(Over)dispersion -> Variable dispersion GLM.
Heteroskedasticity -> Model dispersion.
Zero-inflation -> Add ZIP term.
…
And adjust the model accordingly.
Hurricanes
In https://www.pnas.org/content/111/24/8782 , Jung et al. claim that “Female hurricanes are deadlier than male hurricanes”.
Specifically, they analyze the number of hurricane fatalities, and claim that there is an effect of the femininity of the name on the number of fatalities, correcting for several possible confounders. They interpret the result as causal (including mediators), claiming that giving only male names to hurricanes would considerably reduce death toll.
The data is available in DHARMa.
library (DHARMa)library (mgcv)str (hurricanes)
Classes 'tbl_df', 'tbl' and 'data.frame': 92 obs. of 14 variables:
$ Year : num 1950 1950 1952 1953 1953 ...
$ Name : chr "Easy" "King" "Able" "Barbara" ...
$ MasFem : num 6.78 1.39 3.83 9.83 8.33 ...
$ MinPressure_before : num 958 955 985 987 985 960 954 938 962 987 ...
$ Minpressure_Updated_2014: num 960 955 985 987 985 960 954 938 962 987 ...
$ Gender_MF : num 1 0 0 1 1 1 1 1 1 1 ...
$ Category : num 3 3 1 1 1 3 3 4 3 1 ...
$ alldeaths : num 2 4 3 1 0 60 20 20 0 200 ...
$ NDAM : num 1590 5350 150 58 15 ...
$ Elapsed_Yrs : num 63 63 61 60 60 59 59 59 58 58 ...
$ Source : chr "MWR" "MWR" "MWR" "MWR" ...
$ ZMasFem : num -0.000935 -1.670758 -0.913313 0.945871 0.481075 ...
$ ZMinPressure_A : num -0.356 -0.511 1.038 1.141 1.038 ...
$ ZNDAM : num -0.439 -0.148 -0.55 -0.558 -0.561 ...
Some plots:
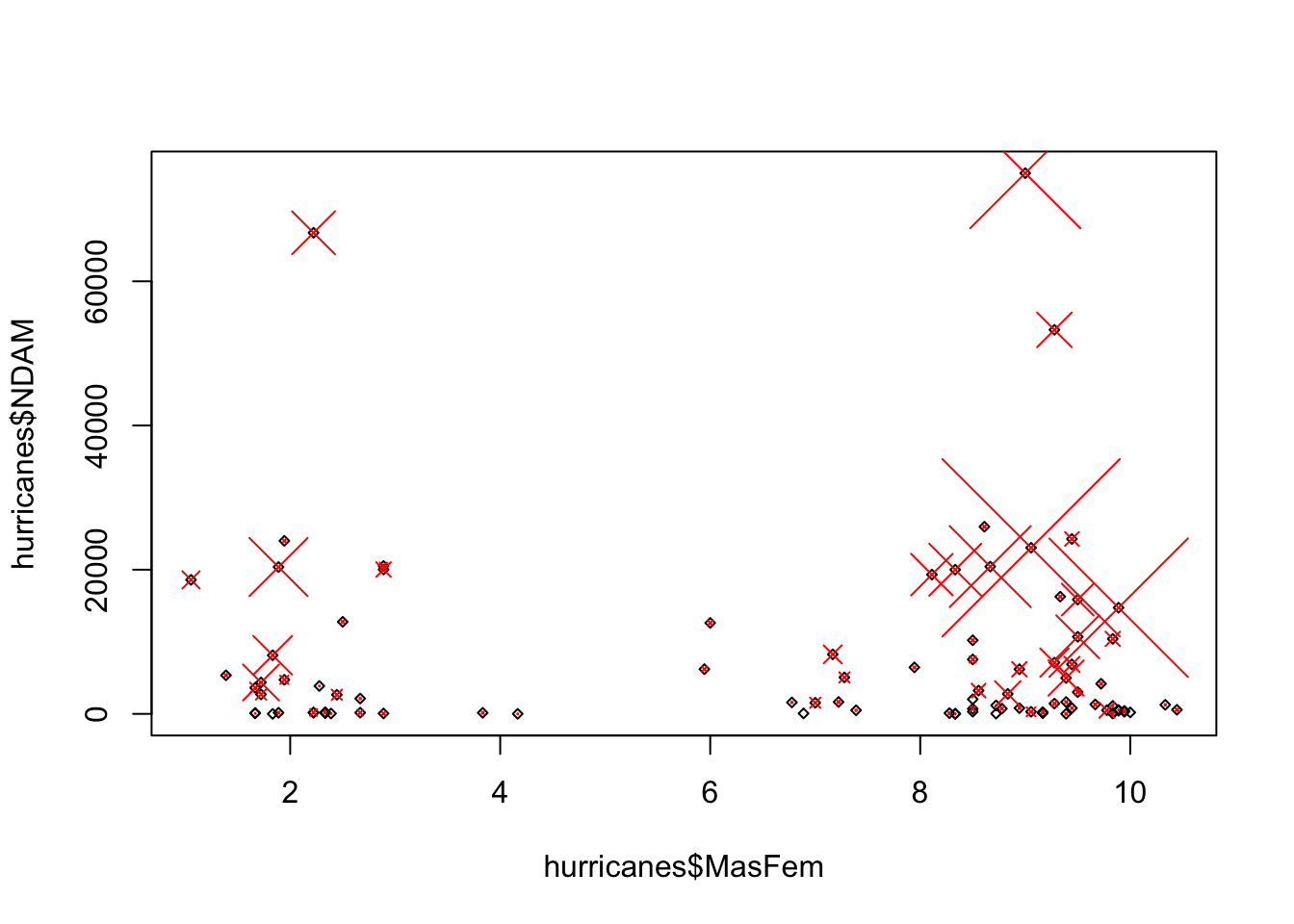
plot (hurricanes$ MasFem, hurricanes$ NDAM, cex = 0.5 , pch = 5 )points (hurricanes$ MasFem, hurricanes$ NDAM, cex = hurricanes$ alldeaths/ 20 ,pch = 4 , col= "red" )
The original model from the paper fits a negative binomial, using mgcv.{R}. I suppose the reason is mainly that glmmTMB was not available at the time, and implementations of the negative binomial, in particular mass::glm.nb and lme4::glmer.nb often had convergence problems.
= gam (alldeaths ~ MasFem * (Minpressure_Updated_2014 + NDAM),data = hurricanes, family = nb, na.action = "na.fail" )summary (originalModelGAM)
Family: Negative Binomial(0.736)
Link function: log
Formula:
alldeaths ~ MasFem * (Minpressure_Updated_2014 + NDAM)
Parametric coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 7.014e+01 2.003e+01 3.502 0.000462 ***
MasFem -5.986e+00 2.529e+00 -2.367 0.017927 *
Minpressure_Updated_2014 -7.008e-02 2.060e-02 -3.402 0.000669 ***
NDAM -3.845e-05 2.945e-05 -1.305 0.191735
MasFem:Minpressure_Updated_2014 6.124e-03 2.603e-03 2.352 0.018656 *
MasFem:NDAM 1.593e-05 3.756e-06 4.242 2.21e-05 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
R-sq.(adj) = -3.61e+03 Deviance explained = 57.4%
-REML = 357.56 Scale est. = 1 n = 92
Tasks:
Confirm that you get the same results as in the paper. It makes sense to translate their model to glmmTMB. Note that the nb parameterization of mgcv corresponds to nbinom2 in glmmTMB. You will get different results when choosing nbinom1
Inspect the fitted model for potential problems, in particular perform a residual analysis of the model, including residuals against all predictors, and improve the model if you find problems.
Forget what they did. Go back to start, do a causal analysis like we did, and do your own model, diagnosing all residual problems that we discussed. Do you think there is an effect of femininity?
This is the model fit by Jung et al., fit with glmmTMB
library (DHARMa)library (glmmTMB)= glmmTMB (alldeaths ~ MasFem* + scale (NDAM)),data = hurricanes, family = nbinom2)summary (m1)
Family: nbinom2 ( log )
Formula: alldeaths ~ MasFem * (Minpressure_Updated_2014 + scale(NDAM))
Data: hurricanes
AIC BIC logLik deviance df.resid
660.7 678.4 -323.4 646.7 85
Dispersion parameter for nbinom2 family (): 0.787
Conditional model:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 69.661590 23.425598 2.974 0.002942 **
MasFem -5.855078 2.716589 -2.155 0.031138 *
Minpressure_Updated_2014 -0.069870 0.024251 -2.881 0.003964 **
scale(NDAM) -0.494094 0.455968 -1.084 0.278536
MasFem:Minpressure_Updated_2014 0.006108 0.002813 2.171 0.029901 *
MasFem:scale(NDAM) 0.205418 0.061956 3.316 0.000915 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Note that in the code that I gave you not all predictors were scaled (and they don’t say if they scaled in the paper), but as we for looking for main effects in the presence of interactions, we should definitely scale to improve the interpretability
= glmmTMB (alldeaths ~ scale (MasFem) * scale (Minpressure_Updated_2014) + scale (NDAM)),data = hurricanes, family = nbinom2)summary (m2)
Family: nbinom2 ( log )
Formula:
alldeaths ~ scale(MasFem) * (scale(Minpressure_Updated_2014) +
scale(NDAM))
Data: hurricanes
AIC BIC logLik deviance df.resid
660.7 678.4 -323.4 646.7 85
Dispersion parameter for nbinom2 family (): 0.787
Conditional model:
Estimate Std. Error z value
(Intercept) 2.5034 0.1231 20.341
scale(MasFem) 0.1237 0.1210 1.022
scale(Minpressure_Updated_2014) -0.5425 0.1603 -3.384
scale(NDAM) 0.8988 0.2190 4.105
scale(MasFem):scale(Minpressure_Updated_2014) 0.3758 0.1731 2.171
scale(MasFem):scale(NDAM) 0.6629 0.1999 3.316
Pr(>|z|)
(Intercept) < 2e-16 ***
scale(MasFem) 0.306923
scale(Minpressure_Updated_2014) 0.000715 ***
scale(NDAM) 4.05e-05 ***
scale(MasFem):scale(Minpressure_Updated_2014) 0.029901 *
scale(MasFem):scale(NDAM) 0.000915 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
now main effect is n.s.; it’s a bit dodgy, but if you read in the main paper, they do not claim a significant main effect, they mainly argue via ANOVA and significance at high values of NDAM, so let’s run an ANOVA:
Analysis of Deviance Table (Type II Wald chisquare tests)
Response: alldeaths
Chisq Df Pr(>Chisq)
scale(MasFem) 1.9495 1 0.1626364
scale(Minpressure_Updated_2014) 7.1285 1 0.0075868 **
scale(NDAM) 14.6100 1 0.0001322 ***
scale(MasFem):scale(Minpressure_Updated_2014) 4.7150 1 0.0299011 *
scale(MasFem):scale(NDAM) 10.9929 1 0.0009146 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
In the ANOVA we see that MasFem still n.s. but interactions, and if you would calculate effect of MasFem at high NDAM, it is significant. Something like that is argued in the paper. We can emulate this by changing NDAM centering to high NDAM, which gives us a p-value for the main effect of MasFem at high values of NDAM
$ highcenteredNDAM = hurricanes$ NDAM - max (hurricanes$ NDAM)= glmmTMB (alldeaths ~ scale (MasFem) * scale (Minpressure_Updated_2014) + highcenteredNDAM),data = hurricanes, family = nbinom2)summary (m3)
Family: nbinom2 ( log )
Formula:
alldeaths ~ scale(MasFem) * (scale(Minpressure_Updated_2014) +
highcenteredNDAM)
Data: hurricanes
AIC BIC logLik deviance df.resid
660.7 678.4 -323.4 646.7 85
Dispersion parameter for nbinom2 family (): 0.787
Conditional model:
Estimate Std. Error z value
(Intercept) 7.210e+00 1.149e+00 6.275
scale(MasFem) 3.595e+00 1.041e+00 3.455
scale(Minpressure_Updated_2014) -5.425e-01 1.603e-01 -3.384
highcenteredNDAM 6.949e-05 1.693e-05 4.105
scale(MasFem):scale(Minpressure_Updated_2014) 3.758e-01 1.731e-01 2.171
scale(MasFem):highcenteredNDAM 5.125e-05 1.546e-05 3.316
Pr(>|z|)
(Intercept) 3.50e-10 ***
scale(MasFem) 0.000551 ***
scale(Minpressure_Updated_2014) 0.000715 ***
highcenteredNDAM 4.05e-05 ***
scale(MasFem):scale(Minpressure_Updated_2014) 0.029904 *
scale(MasFem):highcenteredNDAM 0.000915 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Now we see the significant main effect that they report. Note, hwoever, that the signficant differences is only there for high NDAM, i.e. what we do here is to project the effect of the interaction on the main effect. An alternative to do the same thing would be an effects plot, or to specifically use predict() to calculate differences and CIs at high NDAM values.
Loading required package: carData
lattice theme set by effectsTheme()
See ?effectsTheme for details.
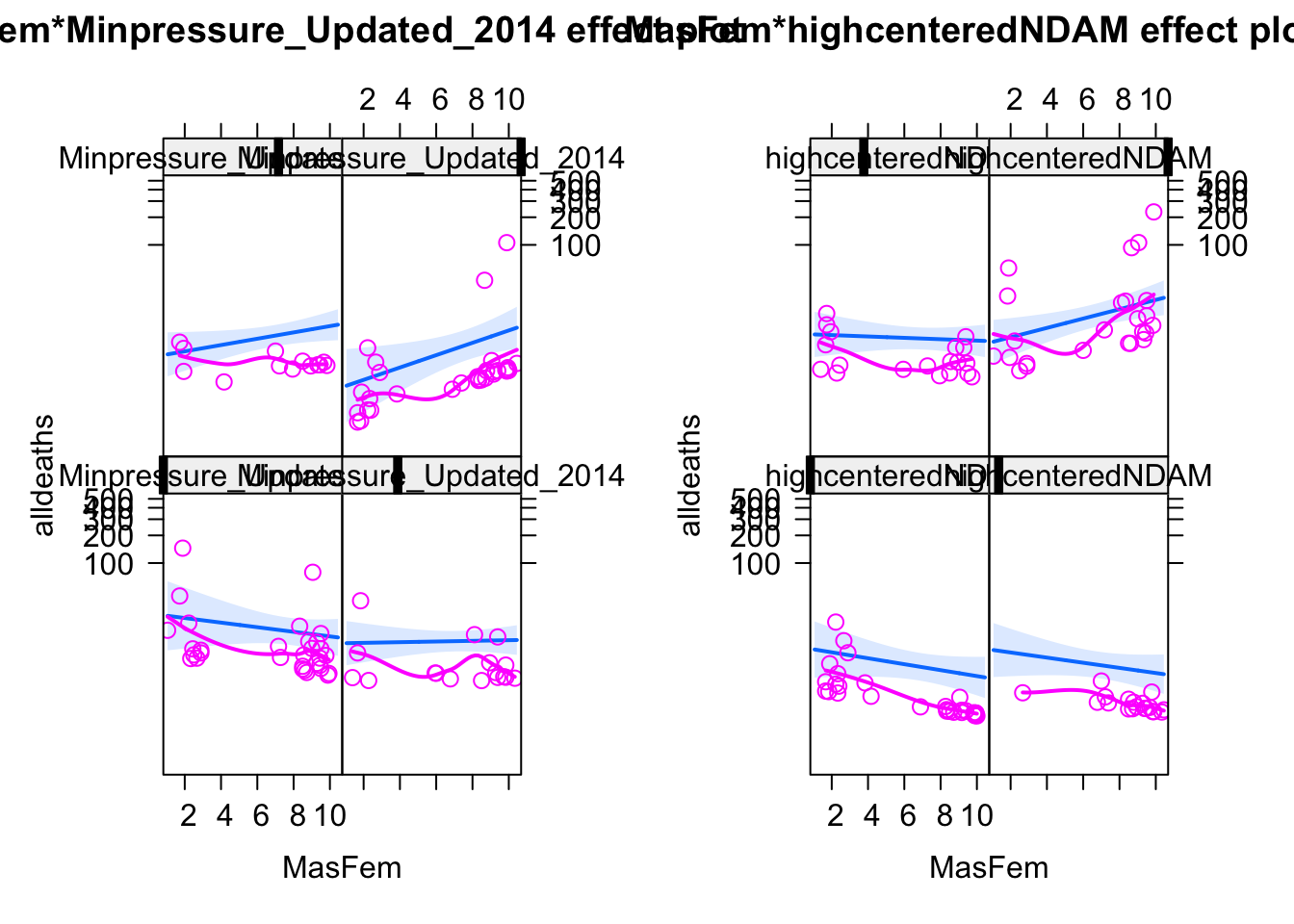
plot (allEffects (m3, partial.residuals = T))
Warning in Effect.glmmTMB(predictors, mod, vcov. = vcov., ...): overriding
variance function for effects/dev.resids: computed variances may be incorrect
Warning in Analyze.model(focal.predictors, mod, xlevels, default.levels, :
the predictors scale(MasFem), scale(Minpressure_Updated_2014) are one-column
matrices that were converted to vectors
Warning in Effect.glmmTMB(predictors, mod, vcov. = vcov., ...): overriding
variance function for effects/dev.resids: computed variances may be incorrect
Warning in Analyze.model(focal.predictors, mod, xlevels, default.levels, :
the predictors scale(MasFem), scale(Minpressure_Updated_2014) are one-column
matrices that were converted to vectors
OK, this means we can replicate the results of the paper, even if concentrating the entire analysis exclusive on high NDAM seems a bit cherry-picking. Another way to phrase the result is that we don’t find a main effect of MasFem. However, to be fair: the current results to say that there is a significant difference at high NDAM, and such a difference, if it existed, would be importat.
But we haven’t done residual checks yet. Let’s do that:
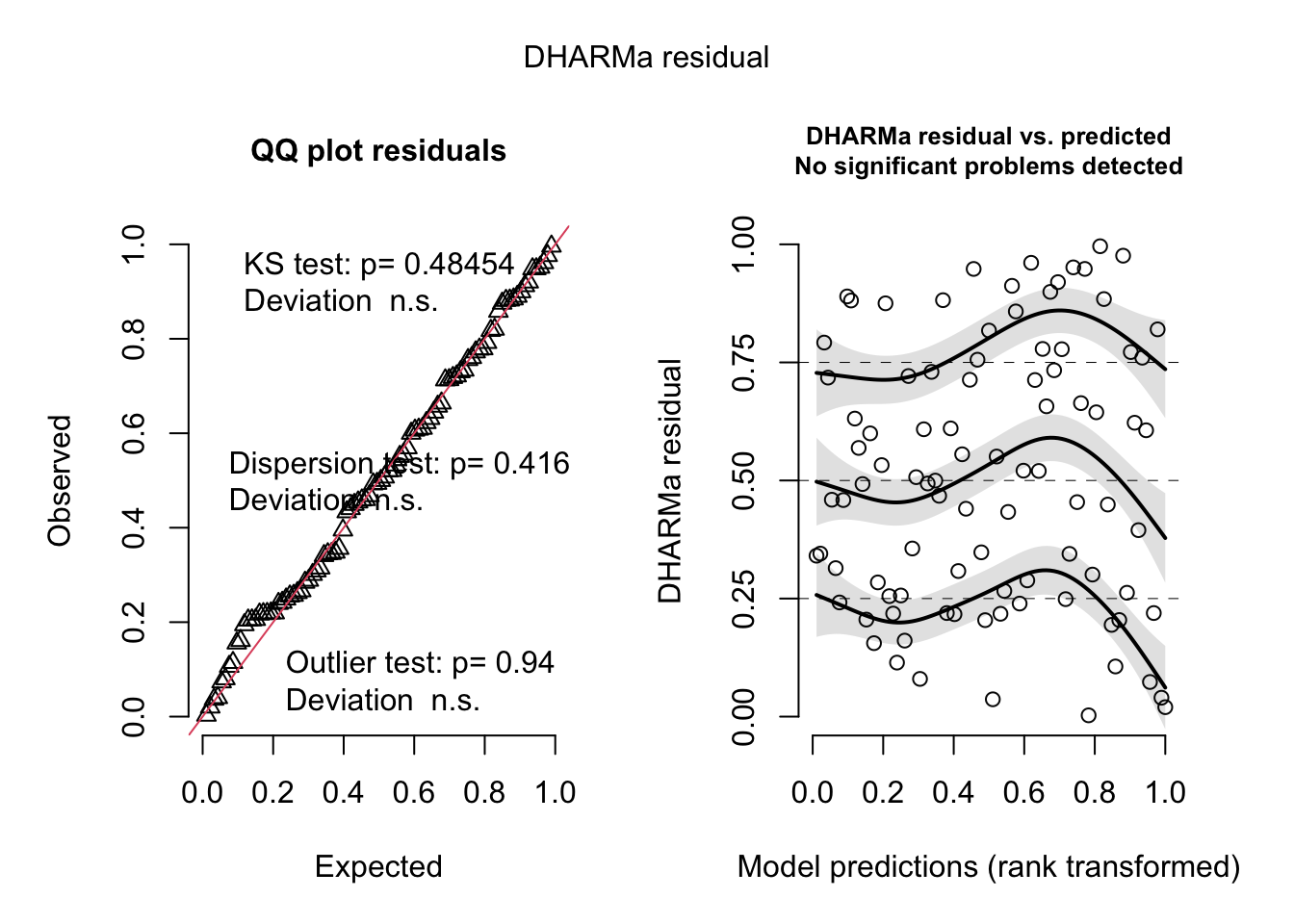
<- simulateResiduals (originalModelGAM)
Registered S3 method overwritten by 'GGally':
method from
+.gg ggplot2
Registered S3 method overwritten by 'mgcViz':
method from
+.gg GGally
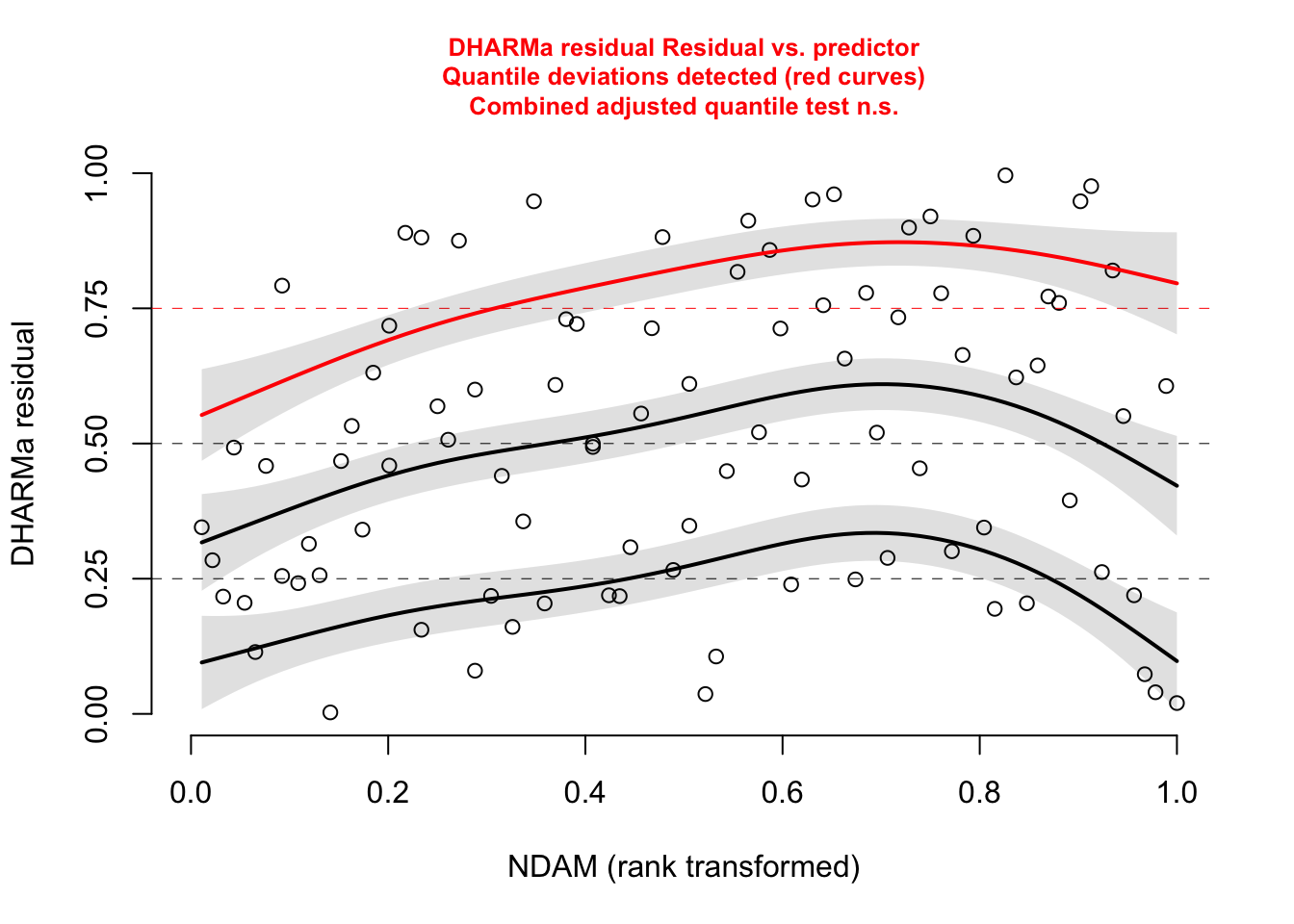
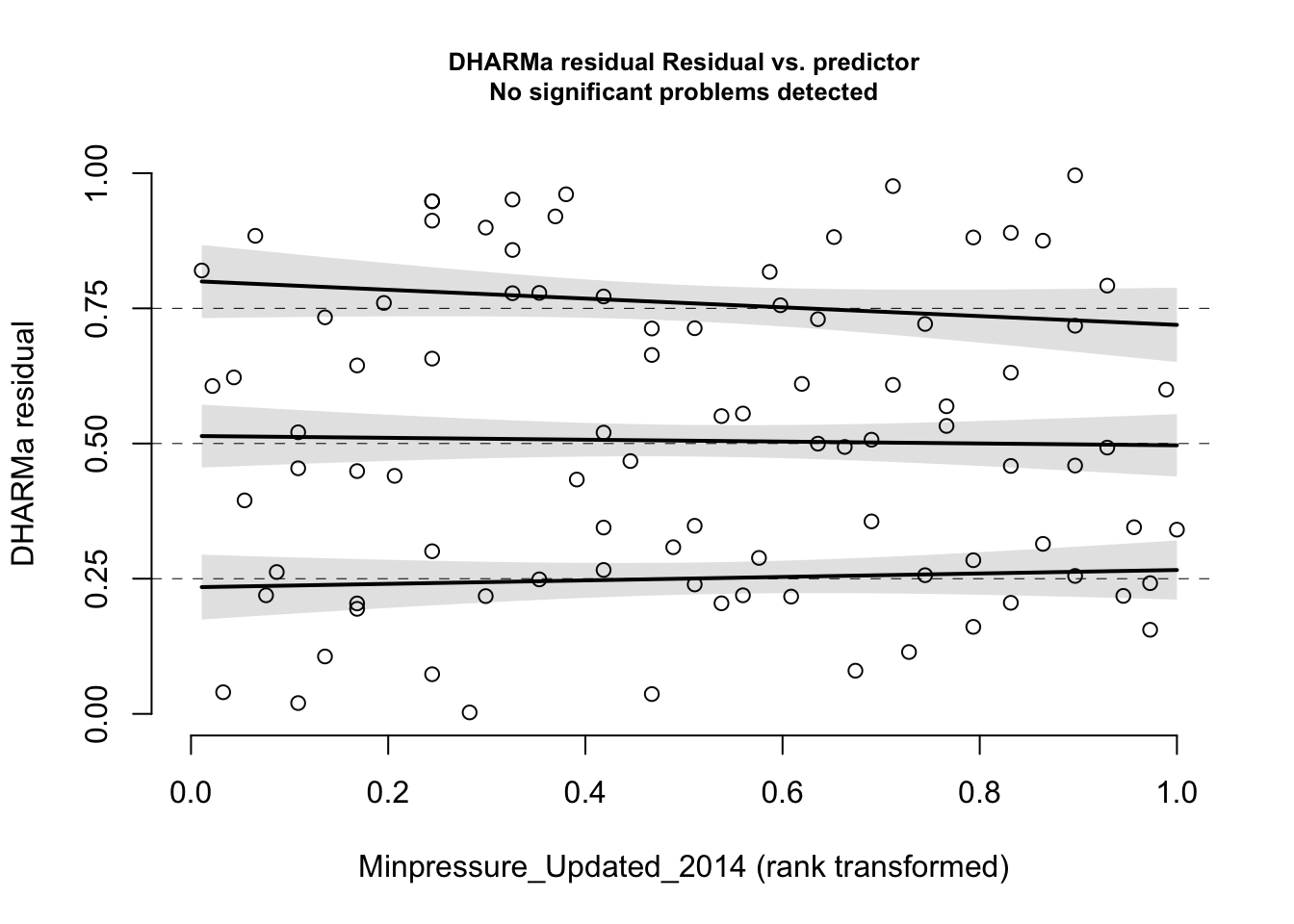
plotResiduals (res, hurricanes$ NDAM)plotResiduals (res, hurricanes$ MasFem)plotResiduals (res, hurricanes$ Minpressure_Updated_2014)
No significant deviation in the general DHARMa plot, but residuals ~ NDAM looks funny, which was also pointed out by Bob O’Hara in a blog post after publication of the paper. Let’s try to correct this - scaling with ^0.2 does a great job:
= glmmTMB (alldeaths ~ scale (MasFem) * scale (Minpressure_Updated_2014) + scale (NDAM^ 0.2 )),data = hurricanes, family = nbinom2)<- simulateResiduals (correctedModel, plot = T)plotResiduals (res, hurricanes$ NDAM)
Family: nbinom2 ( log )
Formula:
alldeaths ~ scale(MasFem) * (scale(Minpressure_Updated_2014) +
scale(NDAM^0.2))
Data: hurricanes
AIC BIC logLik deviance df.resid
630.8 648.5 -308.4 616.8 85
Dispersion parameter for nbinom2 family (): 1.11
Conditional model:
Estimate Std. Error z value
(Intercept) 2.26430 0.10912 20.751
scale(MasFem) 0.05156 0.10695 0.482
scale(Minpressure_Updated_2014) -0.03162 0.18141 -0.174
scale(NDAM^0.2) 1.28961 0.18992 6.790
scale(MasFem):scale(Minpressure_Updated_2014) -0.02410 0.20343 -0.118
scale(MasFem):scale(NDAM^0.2) 0.16045 0.20350 0.788
Pr(>|z|)
(Intercept) < 2e-16 ***
scale(MasFem) 0.630
scale(Minpressure_Updated_2014) 0.862
scale(NDAM^0.2) 1.12e-11 ***
scale(MasFem):scale(Minpressure_Updated_2014) 0.906
scale(MasFem):scale(NDAM^0.2) 0.430
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
:: Anova (correctedModel)
Analysis of Deviance Table (Type II Wald chisquare tests)
Response: alldeaths
Chisq Df Pr(>Chisq)
scale(MasFem) 0.5732 1 0.4490
scale(Minpressure_Updated_2014) 0.1255 1 0.7232
scale(NDAM^0.2) 73.5010 1 <2e-16 ***
scale(MasFem):scale(Minpressure_Updated_2014) 0.0140 1 0.9057
scale(MasFem):scale(NDAM^0.2) 0.6216 1 0.4304
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
All gone, only damage is doing the effect. This wouldn’t change with re-scaling probably, as interactions are n.s.
What if we would have fit our own model? First of all, note that if hurricane names were given randomly, we wouldn’t have to worry about confounders. However, this is not the case, hurricanes were only named randomly after 1978 or so.
plot (MasFem ~ Year, data = hurricanes)
So, we could either take the earlier data out, which would remove half of our data, or we have to worry about confounding with variables that change over time. The most obvious thing would be to take time itself (Year) in the model, to correct for temporal confounding.
Do we need other variables that are not confounders? There is two reasons to add them:
they have strong effects on the response - not adding them could lead to residual problems and increase residual variance, which increases uncertainties and cost power
we want to fit interacts.
I added NDAM to the model, because we saw earlier that it has a strong effect. I think it’s not unreasonable to check for an interaction as well.
As we have several observations per year, a conservative approach would be to add a RE on year. Note that we use year both as a fixed effect (to remove temporal trends) and a random intercept, which is perfectly fine, however.
= glmmTMB (alldeaths ~ scale (MasFem) * scale (NDAM^ 0.2 ) + Year + (1 | Year),data = hurricanes, family = nbinom2)summary (newModel)
Family: nbinom2 ( log )
Formula:
alldeaths ~ scale(MasFem) * scale(NDAM^0.2) + Year + (1 | Year)
Data: hurricanes
AIC BIC logLik deviance df.resid
630.8 648.4 -308.4 616.8 85
Random effects:
Conditional model:
Groups Name Variance Std.Dev.
Year (Intercept) 2.571e-07 0.0005071
Number of obs: 92, groups: Year, 49
Dispersion parameter for nbinom2 family (): 1.11
Conditional model:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -2.542287 12.730846 -0.200 0.842
scale(MasFem) 0.073207 0.119273 0.614 0.539
scale(NDAM^0.2) 1.309624 0.118106 11.089 <2e-16 ***
Year 0.002426 0.006423 0.378 0.706
scale(MasFem):scale(NDAM^0.2) 0.179874 0.117191 1.535 0.125
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
:: Anova (newModel) # nothing regarding MasFem
Analysis of Deviance Table (Type II Wald chisquare tests)
Response: alldeaths
Chisq Df Pr(>Chisq)
scale(MasFem) 0.5563 1 0.4558
scale(NDAM^0.2) 121.9237 1 <2e-16 ***
Year 0.1426 1 0.7057
scale(MasFem):scale(NDAM^0.2) 2.3559 1 0.1248
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
The results remain that there is no effect of MasFem!
Researchers Degrees of Freedom — Skin Color and Red Cards
In 2018 Silberzahn et al. published a “meta analysis” in Advances in Methods and Practices in Psychological Science , where they had provided 29 teams with the same data set to answer one research question: “[W]hether soccer players with dark skin tone are more likely than those with light skin tone to receive red cards from referees ”.
Spoiler : They found that the “[a]nalytic approaches varied widely across the teams, and the estimated effect sizes ranged from 0.89 to 2.93 (Mdn = 1.31) in odds-ratio units”, highlighting that different approaches in data analysis can yield significant variation in the results.
You can find the paper “Many Analysts, One Data Set: Making Transparent How Variations in Analytic Choices Affect Results” at: https://journals.sagepub.com/doi/10.1177/2515245917747646 .
The data is in
library (EcoData)
Task: Do a re-analysis of the data as if you were the 30th team to contribute the results to the meta analysis. You can find the data in the ecodata package, dataset redCards.
Response variable: ‘redCards’ (+‘yellowReds’?).
primary predictors: ‘rater1’, ‘rater2’
Multiple variables, potentially accounting for confounding, offsetting, grouping, … are included in the data.
The rater variable contains ratings of “two independent raters blind to the research question who, based on their profile photo, categorized players on a 5-point scale ranging from (1) very light skin to (5) very dark skin. Make sure that ‘rater1’ and ‘rater2’ are rescaled to the range 0 … 1 as described in the paper (”This variable was rescaled to be bounded by 0 (very light skin) and 1 (very dark skin) prior to the final analysis, to ensure consistency of effect sizes across the teams of analysts. The raw ratings were rescaled to 0, .25, .50, .75, and 1 to create this new scale.”)
When you’re done, have a look at the other modelling teams. Do you understand the models they fit? Note that the results are displayed in terms of odd ratios . Are your results within the range of estimates from the 29 teams in Silberzahn et al. (2018)?
Scouting Ants
Look at the dataset EcoData::scoutingAnts, and find out if there are really scouting Ants in Lasius Niger.
A base model should be:
library (EcoData)= scoutingAnts[scoutingAnts$ first.visit == 0 ,]$ ant_group = as.factor (dat$ ant_group)$ ant_group_main = as.factor (dat$ ant_group_main)<- glm (went.phero ~ ant_group_main, data = dat)summary (fit)
Call:
glm(formula = went.phero ~ ant_group_main, data = dat)
Deviance Residuals:
Min 1Q Median 3Q Max
-0.6986 -0.6624 0.3014 0.3014 0.3376
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.66239 0.02139 30.974 <2e-16 ***
ant_group_mainSecondvisit_1st_to_phero 0.03626 0.02469 1.469 0.142
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for gaussian family taken to be 0.2140339)
Null deviance: 401.35 on 1874 degrees of freedom
Residual deviance: 400.89 on 1873 degrees of freedom
AIC: 2434.5
Number of Fisher Scoring iterations: 2
For me, it made sense to change the contrasts of the possible confounders into something more easily interpretable:
$ directionConst = ifelse (dat$ Treatment %in% c ("LL" , "RR" ), T, F)$ directionPhero = as.factor (ifelse (dat$ Treatment %in% c ("LL" , "RL" ), "left" , "right" ))
Together with the orientation of the Maze, this makes 3 possible directional confounders, and the main predictor (if the Ant went to the pheromone in the first visit).
Adding an RE on colony is logical, and then let’s run the model:
Loading required package: Matrix
Attaching package: 'lme4'
The following object is masked from 'package:nlme':
lmList
<- glmer (went.phero ~ ant_group_main+ directionConst+ directionPhero+ Orientation+ (1 | Colony),family= "binomial" , data= dat)summary (fit1)
Generalized linear mixed model fit by maximum likelihood (Laplace
Approximation) [glmerMod]
Family: binomial ( logit )
Formula: went.phero ~ ant_group_main + directionConst + directionPhero +
Orientation + (1 | Colony)
Data: dat
AIC BIC logLik -2*log(L) df.resid
2192.5 2225.7 -1090.2 2180.5 1869
Scaled residuals:
Min 1Q Median 3Q Max
-4.6870 -1.0451 0.4836 0.6876 1.0318
Random effects:
Groups Name Variance Std.Dev.
Colony (Intercept) 0.9284 0.9636
Number of obs: 1875, groups: Colony, 15
Fixed effects:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 1.9505 0.3330 5.857 4.72e-09 ***
ant_group_mainSecondvisit_1st_to_phero 0.1311 0.1248 1.050 0.29351
directionConstTRUE -1.1465 0.2409 -4.760 1.94e-06 ***
directionPheroright -0.5680 0.1984 -2.863 0.00420 **
Orientationright -0.3493 0.1222 -2.859 0.00425 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Correlation of Fixed Effects:
(Intr) a__S_1 dCTRUE drctnP
ant_g_S_1__ -0.252
drctnCnTRUE -0.413 -0.062
drctnPhrrgh -0.441 -0.001 0.306
Orinttnrght -0.182 -0.094 -0.074 0.085
Surprisingly, we find large effects of the other variables. Because of these large effects, testing for interactions with the experimental treatment as well
<- glmer (went.phero ~ ant_group_main * (+ directionConst+ directionPhero+ Orientation)+ (1 | Colony),family= "binomial" , data= dat)summary (fit2)
Generalized linear mixed model fit by maximum likelihood (Laplace
Approximation) [glmerMod]
Family: binomial ( logit )
Formula: went.phero ~ ant_group_main * (+directionConst + directionPhero +
Orientation) + (1 | Colony)
Data: dat
AIC BIC logLik -2*log(L) df.resid
2131.2 2181.0 -1056.6 2113.2 1866
Scaled residuals:
Min 1Q Median 3Q Max
-6.7899 -0.8936 0.4857 0.5984 1.9020
Random effects:
Groups Name Variance Std.Dev.
Colony (Intercept) 0.9497 0.9745
Number of obs: 1875, groups: Colony, 15
Fixed effects:
Estimate Std. Error
(Intercept) 2.7384 0.3913
ant_group_mainSecondvisit_1st_to_phero -0.7658 0.2763
directionConstTRUE -1.9420 0.3065
directionPheroright -1.8133 0.3007
Orientationright 0.2322 0.2345
ant_group_mainSecondvisit_1st_to_phero:directionConstTRUE 1.0466 0.2638
ant_group_mainSecondvisit_1st_to_phero:directionPheroright 1.4752 0.2664
ant_group_mainSecondvisit_1st_to_phero:Orientationright -0.8261 0.2668
z value Pr(>|z|)
(Intercept) 6.999 2.58e-12 ***
ant_group_mainSecondvisit_1st_to_phero -2.772 0.00557 **
directionConstTRUE -6.337 2.34e-10 ***
directionPheroright -6.030 1.64e-09 ***
Orientationright 0.990 0.32202
ant_group_mainSecondvisit_1st_to_phero:directionConstTRUE 3.967 7.28e-05 ***
ant_group_mainSecondvisit_1st_to_phero:directionPheroright 5.538 3.07e-08 ***
ant_group_mainSecondvisit_1st_to_phero:Orientationright -3.097 0.00196 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Correlation of Fixed Effects:
(Intr) an__S_1__ dCTRUE drctnP Ornttn a__S_1__:C a__S_1__:P
ant_g_S_1__ -0.532
drctnCnTRUE -0.433 0.241
drctnPhrrgh -0.512 0.369 0.186
Orinttnrght -0.282 0.428 -0.100 0.008
a__S_1__:CT 0.217 -0.443 -0.592 0.043 0.015
an__S_1__:P 0.342 -0.528 0.007 -0.733 -0.048 -0.072
an__S_1__:O 0.225 -0.526 0.089 0.034 -0.845 -0.021 0.046
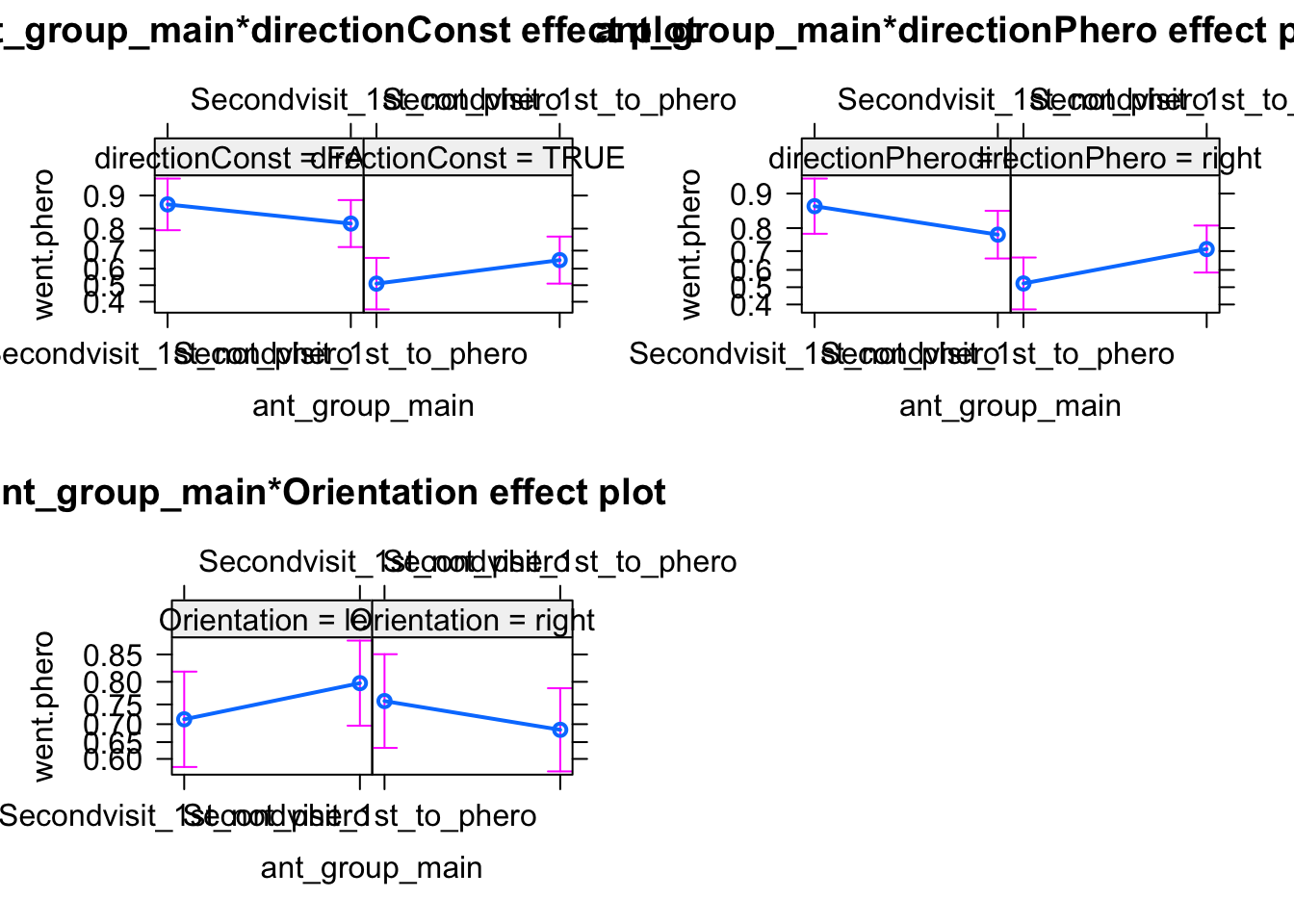
Here we find now that ther is an interaction with the main predictor, and there could be effects. We can also look at this visually.
The results are difficult to interpret. I would think that there was some bias in the experiment, which led to an effect of the Maze direction, which then create a spill-over to the other (and in particular the main) predictors.
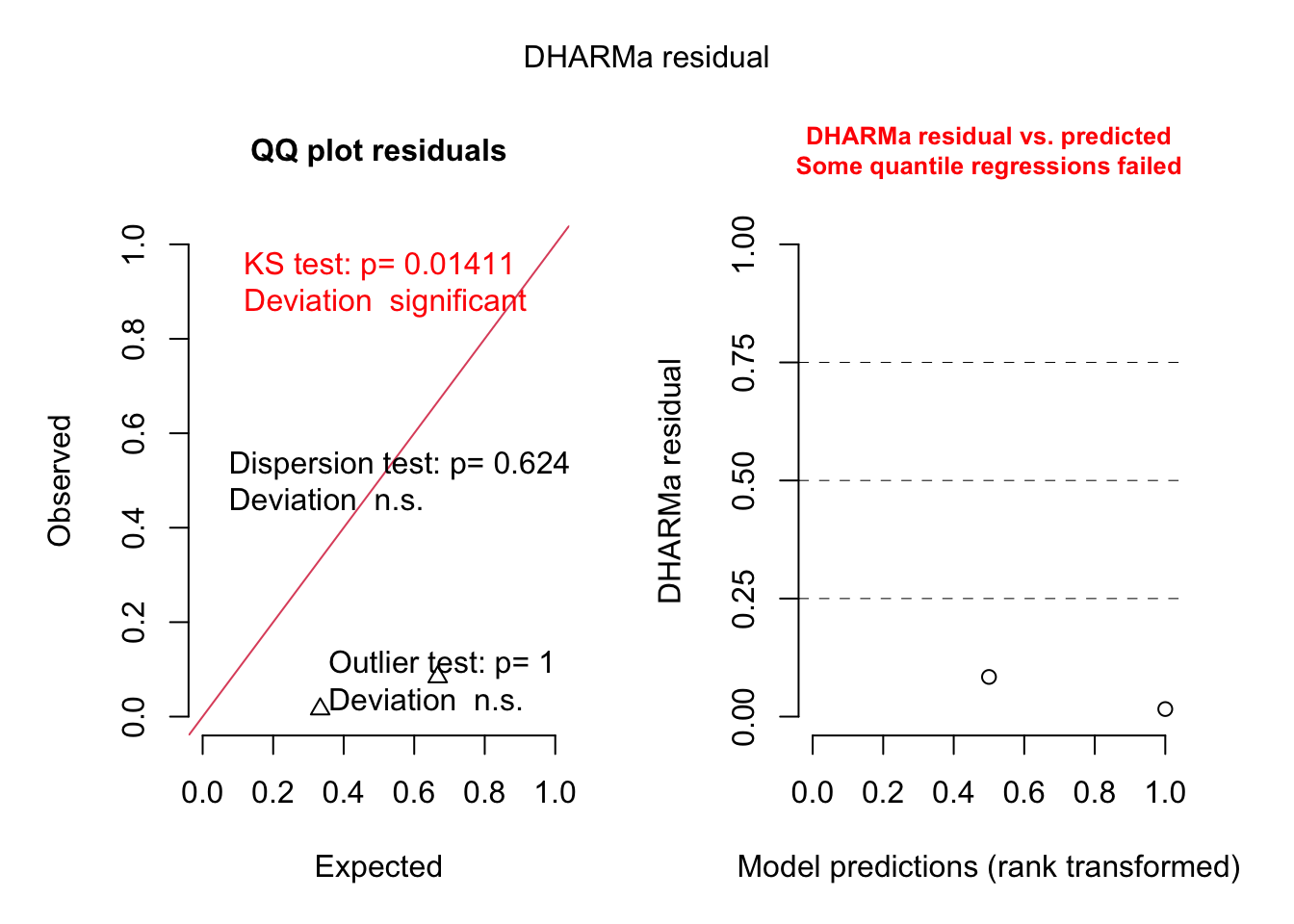
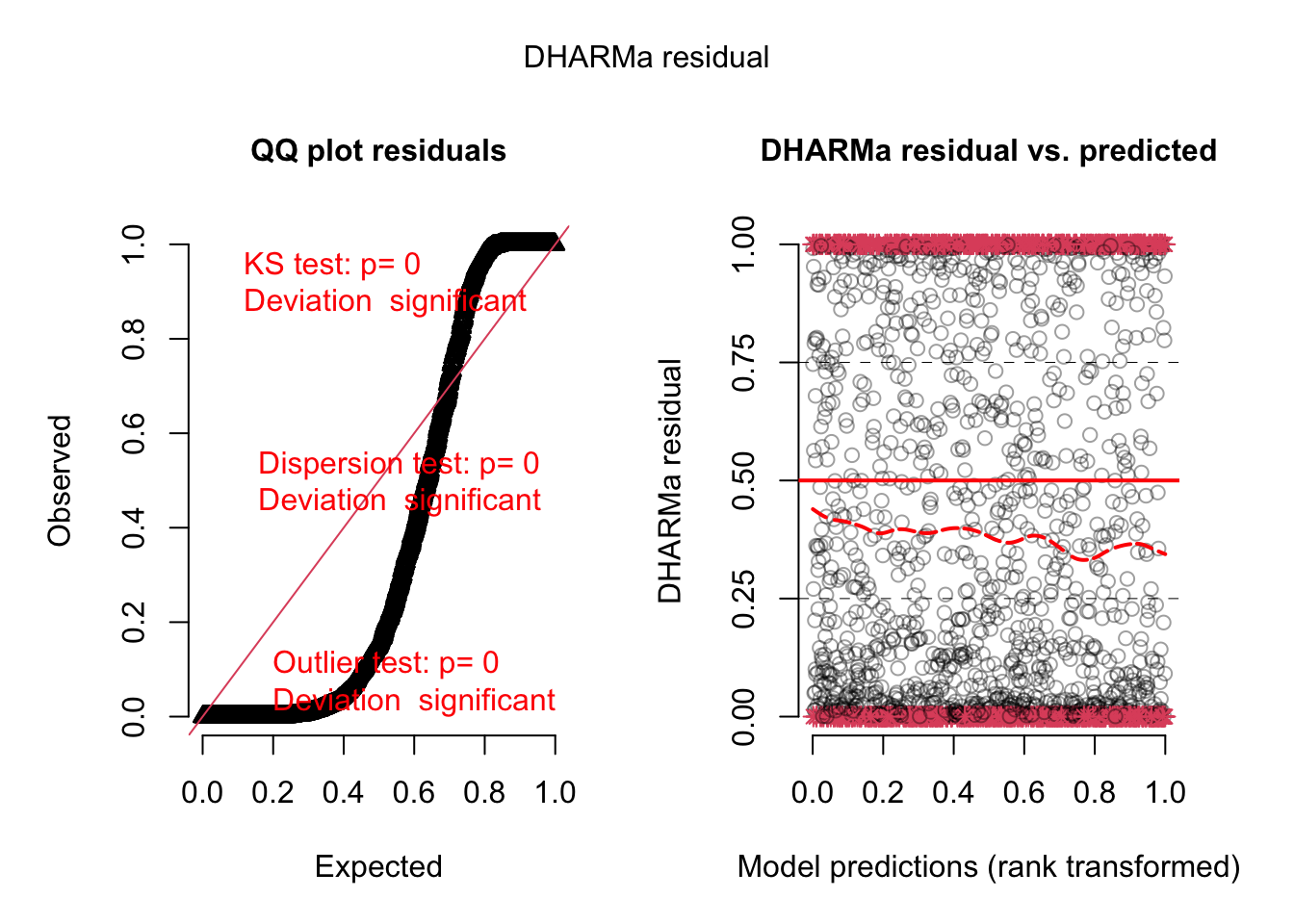
For our education, we can also look at the residual plots. I will use m1, because there was a misfit:
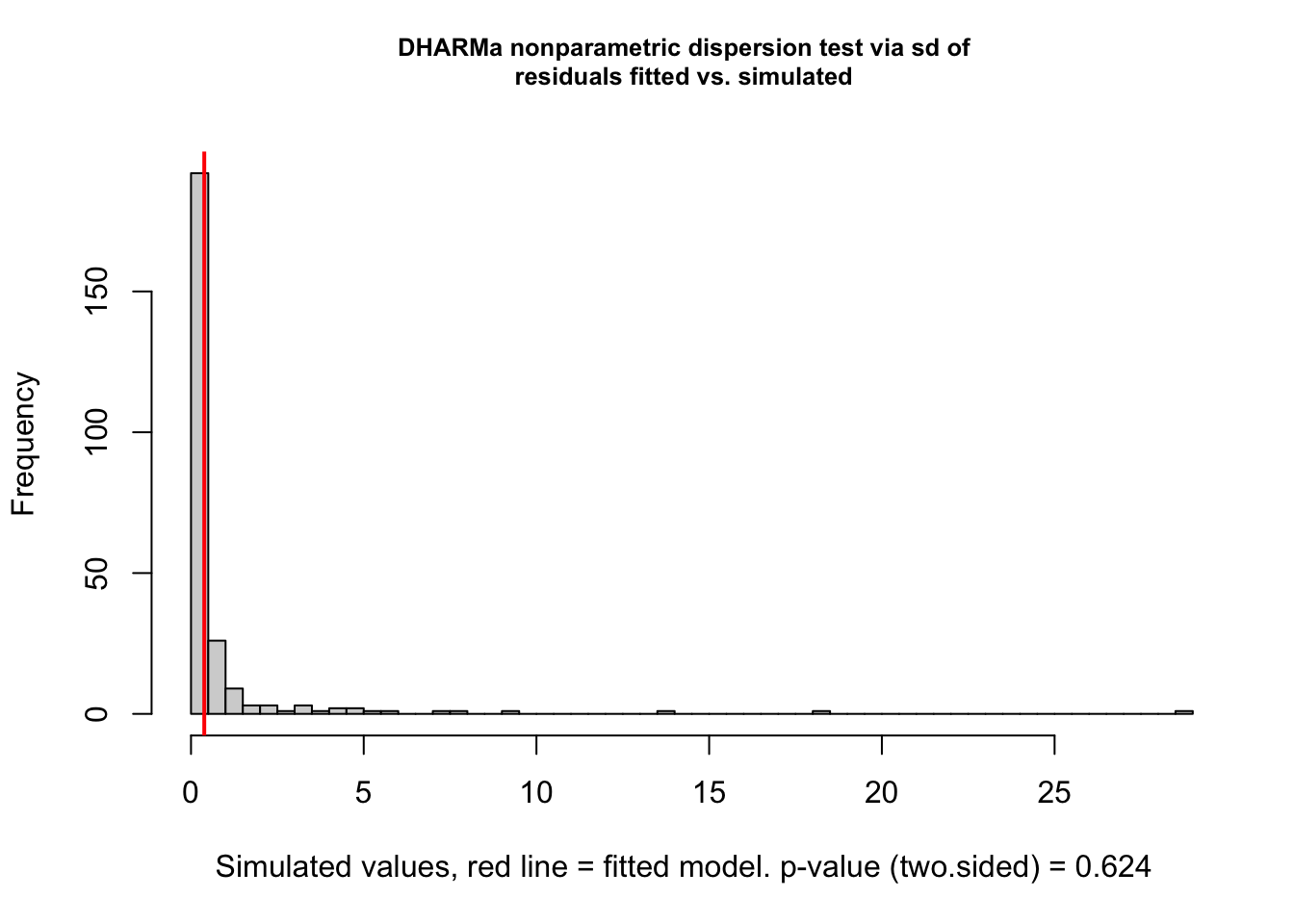
<- simulateResiduals (m1, plot = T)
As we would significant interactions, we would probably see something if we plot residuals against predictors or their interactions, but I want to show you something else:
We will not see dispersion problems in a 0/1 binomial, but actually, this is a k/n binomial, just that the data are not prepared as such.
Either way, in DHARMa, you can aggregate residuals by a grouping variable.
<- recalculateResiduals (res, group = dat$ Colony)
Now, we essentially check k/n data, and we see that there is overdispersion, which is caused by the misfit.
Warning in smooth.construct.tp.smooth.spec(object, dk$data, dk$knots): basis dimension, k, increased to minimum possible
DHARMa: qgam was unable to calculate quantile regression for quantile 0.25. Possibly to few (unique) data points / predictions. The quantile will be ommited in plots and significance calculations.
Warning in smooth.construct.tp.smooth.spec(object, dk$data, dk$knots): basis dimension, k, increased to minimum possible
DHARMa: qgam was unable to calculate quantile regression for quantile 0.5. Possibly to few (unique) data points / predictions. The quantile will be ommited in plots and significance calculations.
Warning in smooth.construct.tp.smooth.spec(object, dk$data, dk$knots): basis dimension, k, increased to minimum possible
DHARMa: qgam was unable to calculate quantile regression for quantile 0.75. Possibly to few (unique) data points / predictions. The quantile will be ommited in plots and significance calculations.
DHARMa nonparametric dispersion test via sd of residuals fitted vs.
simulated
data: simulationOutput
dispersion = 0.47405, p-value = 0.624
alternative hypothesis: two.sided
Let’s do the same for model 2, which included the interactions.
<- simulateResiduals (m2, plot = T)<- recalculateResiduals (res, group = dat$ Colony)plot (res2)
Warning in smooth.construct.tp.smooth.spec(object, dk$data, dk$knots): basis dimension, k, increased to minimum possible
DHARMa: qgam was unable to calculate quantile regression for quantile 0.25. Possibly to few (unique) data points / predictions. The quantile will be ommited in plots and significance calculations.
Warning in smooth.construct.tp.smooth.spec(object, dk$data, dk$knots): basis dimension, k, increased to minimum possible
DHARMa: qgam was unable to calculate quantile regression for quantile 0.5. Possibly to few (unique) data points / predictions. The quantile will be ommited in plots and significance calculations.
Warning in smooth.construct.tp.smooth.spec(object, dk$data, dk$knots): basis dimension, k, increased to minimum possible
DHARMa: qgam was unable to calculate quantile regression for quantile 0.75. Possibly to few (unique) data points / predictions. The quantile will be ommited in plots and significance calculations.
DHARMa nonparametric dispersion test via sd of residuals fitted vs.
simulated
data: simulationOutput
dispersion = 0.47405, p-value = 0.624
alternative hypothesis: two.sided
Which largely removes the problem!
Owls
Look at the Owl data set in the glmmTMB package. The initial hypothesis is
library (glmmTMB)= glm (SiblingNegotiation ~ FoodTreatment* SexParent + offset (log (BroodSize)),data = Owls , family = poisson)= simulateResiduals (m1)plot (res)
The offset is a special command that can be used in all regression models. It means that we include an effect with effect size 1.
The offset has a special importance in models with a log link function, because with these models, we have y = exp(x …), so if you do y = exp(x + log(BroodSize) ) and use exp rules, this is y = exp(x) * exp(log(BroodSize)) = y = exp(x) * BroodSize, so this makes the response proportional to BroodSize. This trick is often used in log link GLMs to make the response proportional to Area, Sampling effort, etc.
Task: try to improve the model with everything we have discussed so far.
= glmmTMB:: glmmTMB (SiblingNegotiation ~ FoodTreatment * SexParent+ (1 | Nest) + offset (log (BroodSize)), data = Owls , family = nbinom1,dispformula = ~ FoodTreatment + SexParent,ziformula = ~ FoodTreatment + SexParent)summary (m1)
Family: nbinom1 ( log )
Formula:
SiblingNegotiation ~ FoodTreatment * SexParent + (1 | Nest) +
offset(log(BroodSize))
Zero inflation: ~FoodTreatment + SexParent
Dispersion: ~FoodTreatment + SexParent
Data: Owls
AIC BIC logLik deviance df.resid
3354.6 3402.9 -1666.3 3332.6 588
Random effects:
Conditional model:
Groups Name Variance Std.Dev.
Nest (Intercept) 0.0876 0.296
Number of obs: 599, groups: Nest, 27
Conditional model:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 0.80028 0.09736 8.220 < 2e-16 ***
FoodTreatmentSatiated -0.46893 0.16760 -2.798 0.00514 **
SexParentMale -0.09127 0.09247 -0.987 0.32363
FoodTreatmentSatiated:SexParentMale 0.13087 0.19028 0.688 0.49158
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Zero-inflation model:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -1.9132 0.3269 -5.853 4.84e-09 ***
FoodTreatmentSatiated 1.0564 0.4072 2.594 0.00948 **
SexParentMale -0.4688 0.3659 -1.281 0.20012
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Dispersion model:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 1.2122 0.2214 5.475 4.37e-08 ***
FoodTreatmentSatiated 0.7978 0.2732 2.920 0.0035 **
SexParentMale -0.1540 0.2399 -0.642 0.5209
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
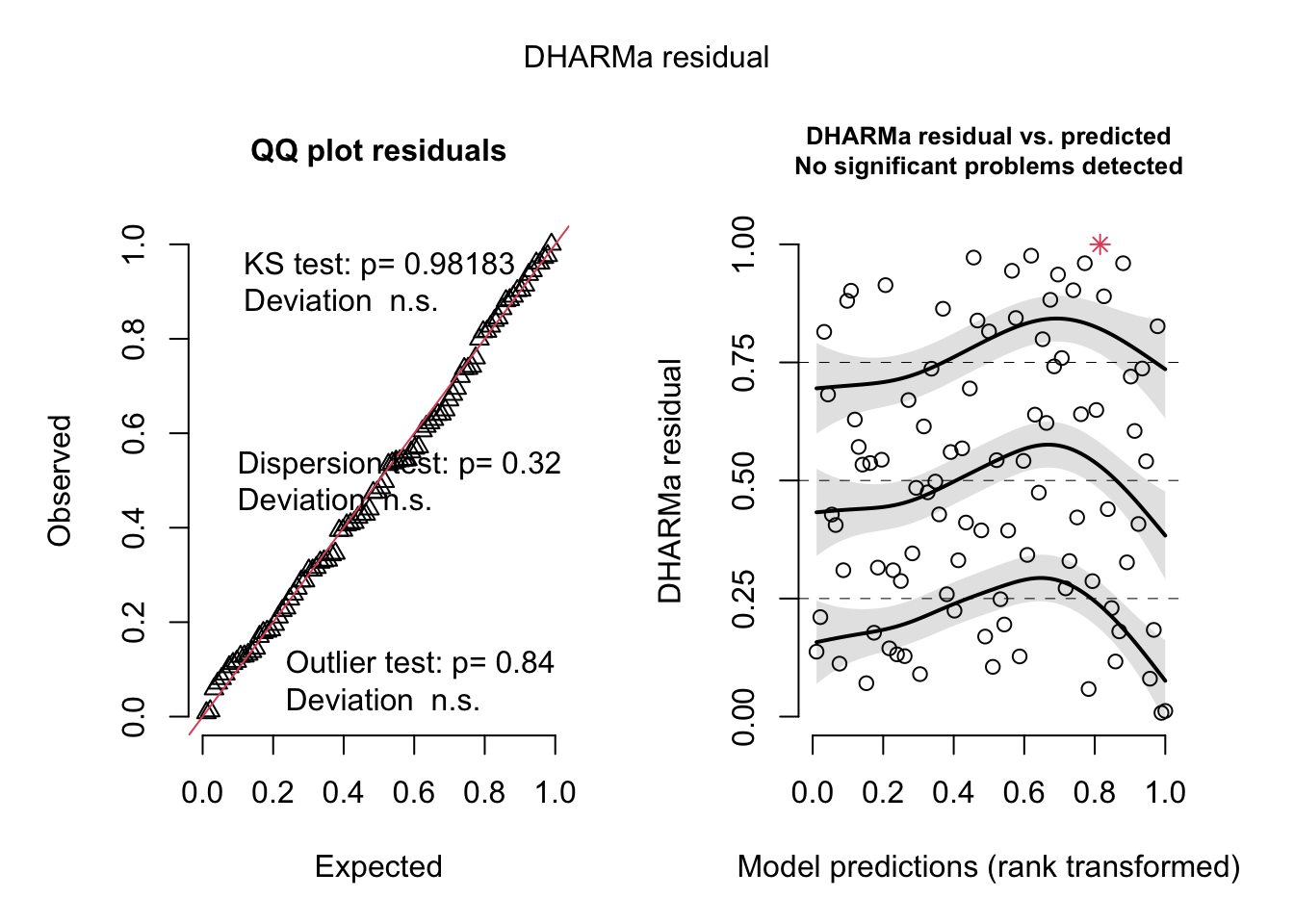
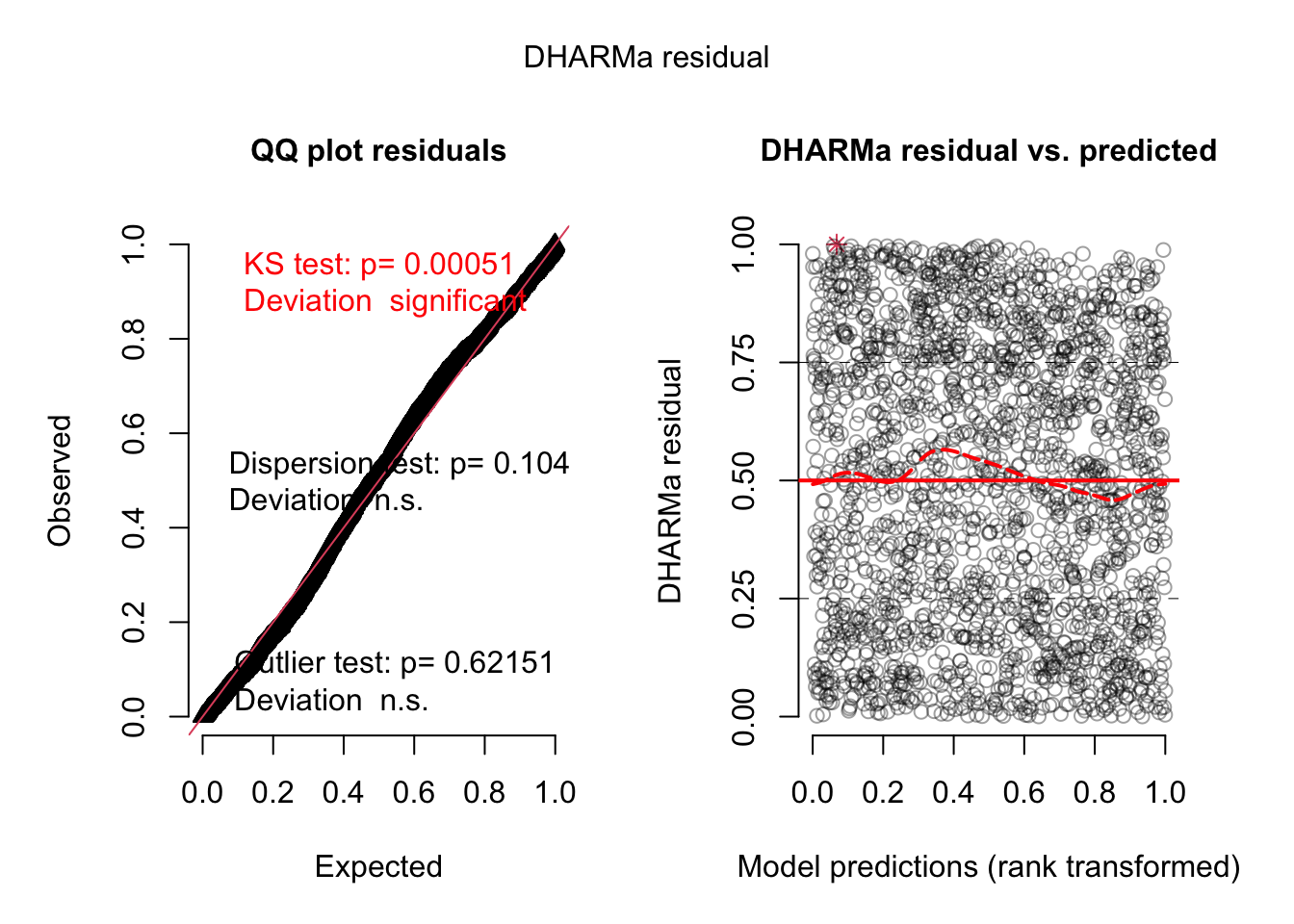
= simulateResiduals (m1, plot = T)
DHARMa nonparametric dispersion test via sd of residuals fitted vs.
simulated
data: simulationOutput
dispersion = 0.78311, p-value = 0.104
alternative hypothesis: two.sided
DHARMa zero-inflation test via comparison to expected zeros with
simulation under H0 = fitted model
data: simulationOutput
ratioObsSim = 1.0465, p-value = 0.608
alternative hypothesis: two.sided
This is not adding dispersion and zero-inflation yet, just to show how such a model could be fit with brms
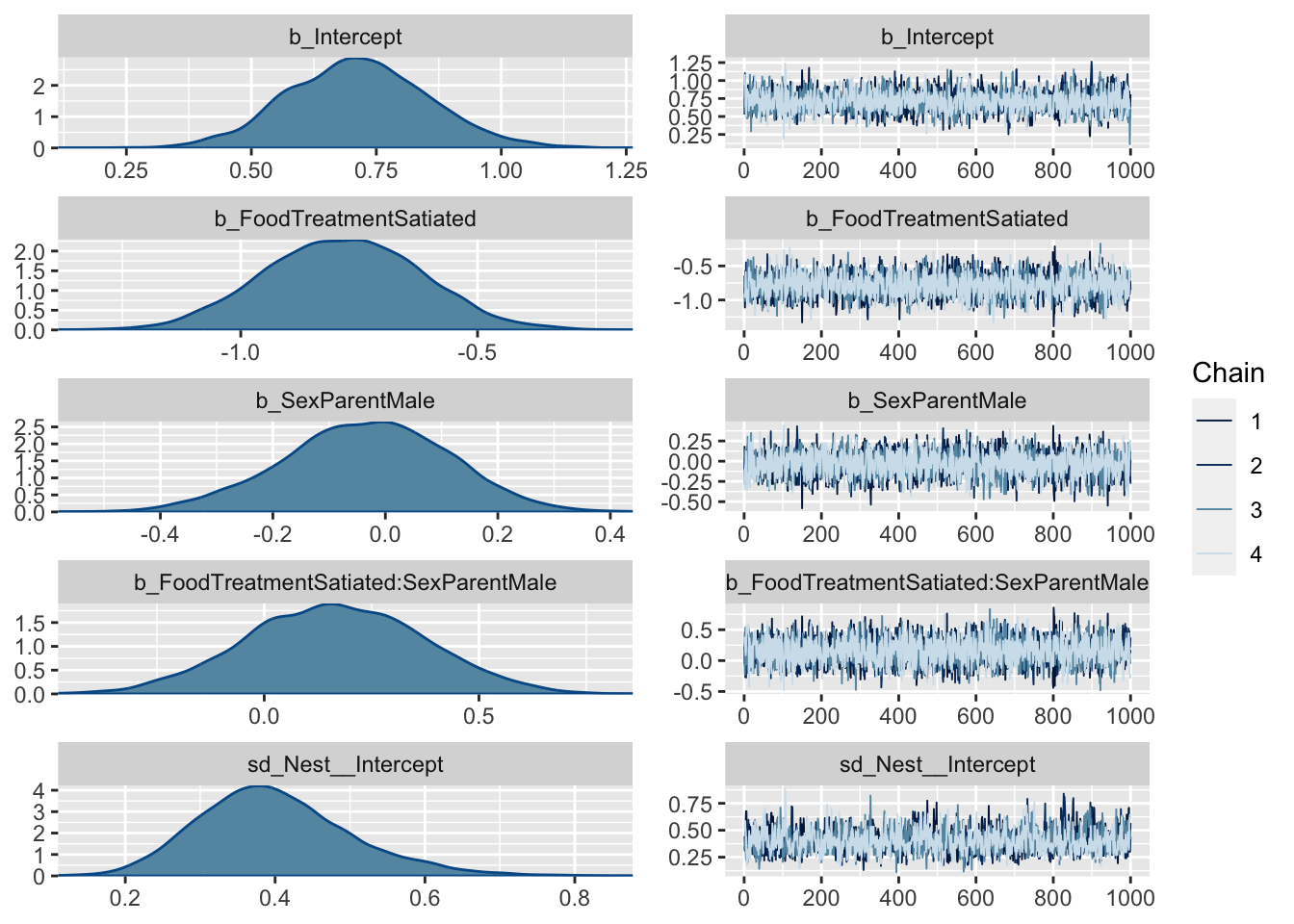
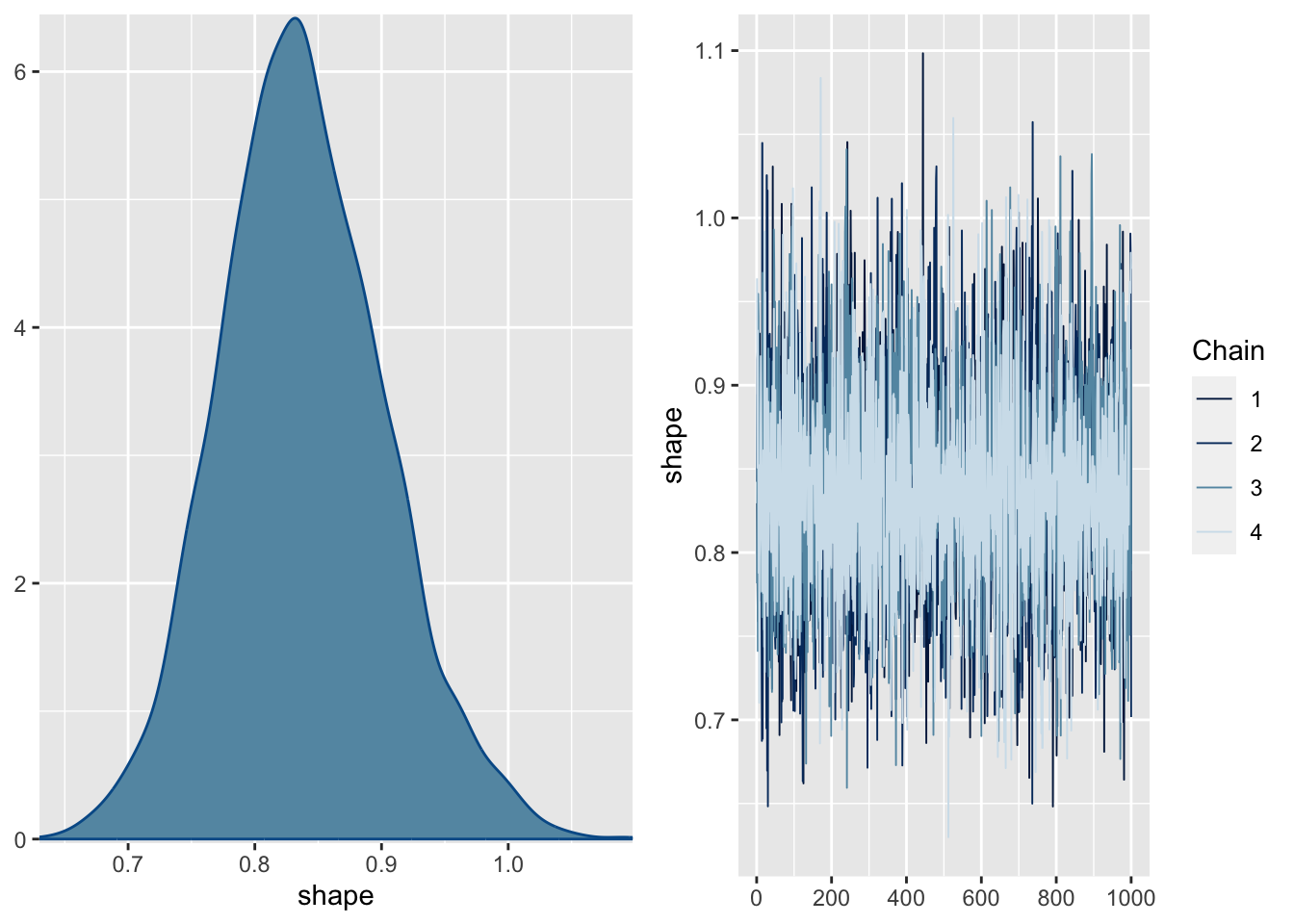
library (brms)= brms:: brm (SiblingNegotiation ~ FoodTreatment * SexParent+ (1 | Nest) + offset (log (BroodSize)), data = Owls , family = negbinomial)
SAMPLING FOR MODEL 'anon_model' NOW (CHAIN 1).
Chain 1:
Chain 1: Gradient evaluation took 0.000209 seconds
Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 2.09 seconds.
Chain 1: Adjust your expectations accordingly!
Chain 1:
Chain 1:
Chain 1: Iteration: 1 / 2000 [ 0%] (Warmup)
Chain 1: Iteration: 200 / 2000 [ 10%] (Warmup)
Chain 1: Iteration: 400 / 2000 [ 20%] (Warmup)
Chain 1: Iteration: 600 / 2000 [ 30%] (Warmup)
Chain 1: Iteration: 800 / 2000 [ 40%] (Warmup)
Chain 1: Iteration: 1000 / 2000 [ 50%] (Warmup)
Chain 1: Iteration: 1001 / 2000 [ 50%] (Sampling)
Chain 1: Iteration: 1200 / 2000 [ 60%] (Sampling)
Chain 1: Iteration: 1400 / 2000 [ 70%] (Sampling)
Chain 1: Iteration: 1600 / 2000 [ 80%] (Sampling)
Chain 1: Iteration: 1800 / 2000 [ 90%] (Sampling)
Chain 1: Iteration: 2000 / 2000 [100%] (Sampling)
Chain 1:
Chain 1: Elapsed Time: 1.328 seconds (Warm-up)
Chain 1: 1.159 seconds (Sampling)
Chain 1: 2.487 seconds (Total)
Chain 1:
SAMPLING FOR MODEL 'anon_model' NOW (CHAIN 2).
Chain 2:
Chain 2: Gradient evaluation took 9.1e-05 seconds
Chain 2: 1000 transitions using 10 leapfrog steps per transition would take 0.91 seconds.
Chain 2: Adjust your expectations accordingly!
Chain 2:
Chain 2:
Chain 2: Iteration: 1 / 2000 [ 0%] (Warmup)
Chain 2: Iteration: 200 / 2000 [ 10%] (Warmup)
Chain 2: Iteration: 400 / 2000 [ 20%] (Warmup)
Chain 2: Iteration: 600 / 2000 [ 30%] (Warmup)
Chain 2: Iteration: 800 / 2000 [ 40%] (Warmup)
Chain 2: Iteration: 1000 / 2000 [ 50%] (Warmup)
Chain 2: Iteration: 1001 / 2000 [ 50%] (Sampling)
Chain 2: Iteration: 1200 / 2000 [ 60%] (Sampling)
Chain 2: Iteration: 1400 / 2000 [ 70%] (Sampling)
Chain 2: Iteration: 1600 / 2000 [ 80%] (Sampling)
Chain 2: Iteration: 1800 / 2000 [ 90%] (Sampling)
Chain 2: Iteration: 2000 / 2000 [100%] (Sampling)
Chain 2:
Chain 2: Elapsed Time: 1.448 seconds (Warm-up)
Chain 2: 1.292 seconds (Sampling)
Chain 2: 2.74 seconds (Total)
Chain 2:
SAMPLING FOR MODEL 'anon_model' NOW (CHAIN 3).
Chain 3:
Chain 3: Gradient evaluation took 9.1e-05 seconds
Chain 3: 1000 transitions using 10 leapfrog steps per transition would take 0.91 seconds.
Chain 3: Adjust your expectations accordingly!
Chain 3:
Chain 3:
Chain 3: Iteration: 1 / 2000 [ 0%] (Warmup)
Chain 3: Iteration: 200 / 2000 [ 10%] (Warmup)
Chain 3: Iteration: 400 / 2000 [ 20%] (Warmup)
Chain 3: Iteration: 600 / 2000 [ 30%] (Warmup)
Chain 3: Iteration: 800 / 2000 [ 40%] (Warmup)
Chain 3: Iteration: 1000 / 2000 [ 50%] (Warmup)
Chain 3: Iteration: 1001 / 2000 [ 50%] (Sampling)
Chain 3: Iteration: 1200 / 2000 [ 60%] (Sampling)
Chain 3: Iteration: 1400 / 2000 [ 70%] (Sampling)
Chain 3: Iteration: 1600 / 2000 [ 80%] (Sampling)
Chain 3: Iteration: 1800 / 2000 [ 90%] (Sampling)
Chain 3: Iteration: 2000 / 2000 [100%] (Sampling)
Chain 3:
Chain 3: Elapsed Time: 1.303 seconds (Warm-up)
Chain 3: 1.056 seconds (Sampling)
Chain 3: 2.359 seconds (Total)
Chain 3:
SAMPLING FOR MODEL 'anon_model' NOW (CHAIN 4).
Chain 4:
Chain 4: Gradient evaluation took 8.8e-05 seconds
Chain 4: 1000 transitions using 10 leapfrog steps per transition would take 0.88 seconds.
Chain 4: Adjust your expectations accordingly!
Chain 4:
Chain 4:
Chain 4: Iteration: 1 / 2000 [ 0%] (Warmup)
Chain 4: Iteration: 200 / 2000 [ 10%] (Warmup)
Chain 4: Iteration: 400 / 2000 [ 20%] (Warmup)
Chain 4: Iteration: 600 / 2000 [ 30%] (Warmup)
Chain 4: Iteration: 800 / 2000 [ 40%] (Warmup)
Chain 4: Iteration: 1000 / 2000 [ 50%] (Warmup)
Chain 4: Iteration: 1001 / 2000 [ 50%] (Sampling)
Chain 4: Iteration: 1200 / 2000 [ 60%] (Sampling)
Chain 4: Iteration: 1400 / 2000 [ 70%] (Sampling)
Chain 4: Iteration: 1600 / 2000 [ 80%] (Sampling)
Chain 4: Iteration: 1800 / 2000 [ 90%] (Sampling)
Chain 4: Iteration: 2000 / 2000 [100%] (Sampling)
Chain 4:
Chain 4: Elapsed Time: 1.286 seconds (Warm-up)
Chain 4: 0.931 seconds (Sampling)
Chain 4: 2.217 seconds (Total)
Chain 4:
Family: negbinomial
Links: mu = log; shape = identity
Formula: SiblingNegotiation ~ FoodTreatment * SexParent + (1 | Nest) + offset(log(BroodSize))
Data: Owls (Number of observations: 599)
Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
total post-warmup draws = 4000
Group-Level Effects:
~Nest (Number of levels: 27)
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
sd(Intercept) 0.39 0.10 0.21 0.61 1.00 1374 2113
Population-Level Effects:
Estimate Est.Error l-95% CI u-95% CI Rhat
Intercept 0.72 0.14 0.45 0.99 1.00
FoodTreatmentSatiated -0.78 0.16 -1.09 -0.45 1.00
SexParentMale -0.03 0.14 -0.32 0.25 1.00
FoodTreatmentSatiated:SexParentMale 0.16 0.21 -0.25 0.56 1.00
Bulk_ESS Tail_ESS
Intercept 2668 2826
FoodTreatmentSatiated 3341 2963
SexParentMale 3450 3277
FoodTreatmentSatiated:SexParentMale 3242 2710
Family Specific Parameters:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
shape 0.84 0.07 0.72 0.98 1.00 5466 2991
Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
and Tail_ESS are effective sample size measures, and Rhat is the potential
scale reduction factor on split chains (at convergence, Rhat = 1).
Snails
library (EcoData)library (glmmTMB)library (lme4)library (DHARMa)library (tidyverse):: snails
Look at the Snails data set in the EcoData package, and find out which environmental and/or seasonal predictors i) explain the total abundance and ii) the infection rate of the three species.
The snails data set in the EcoData package includes observations on the distribution of freshwater snails and their infection rates ( schistosomiasis (a parasit)).
The first scientific question is that their adbundance depends on the water conditions. The second scientific question is that their infection rate depends on the water conditions and seasonsal factors
The data also contains data on other environmental (and seasonal factors). You should consider if it is useful to add them to the analysis.
Species: BP_tot, BF_tot, BT_tot
Number of infected individuals: BP_pos_tot, BF_pos_tot, BT_pos_tot
Total abundances of BP species: Bulinus_tot
Total number of infected in BP species: Bulinus_pos_tot
Tasks:
Model the summed total abundance of the three species (Bulinus_tot)
Model the infection rate of all three species (Bulinuts_pos_tot) (k/n binomial)
Optional: Model the species individually (BP_tot, BF_tot, BT_tot)
Optional: Fit a multivariate (joint) species distribution model
Prepare+scale data:
library (lme4)library (glmmTMB)library (DHARMa)= EcoData:: snails$ sTemp_Water = scale (data$ Temp_Water)$ spH = scale (data$ pH)$ swater_speed_ms = scale (data$ water_speed_ms)$ swater_depth = scale (data$ water_depth)$ sCond = scale (data$ Cond)$ swmo_prec = scale (data$ wmo_prec)$ syear = scale (data$ year)$ sLat = scale (data$ Latitude)$ sLon = scale (data$ Longitude)$ sTemp_Air = scale (data$ Temp_Air)# Let's remove NAs beforehand: = rownames (model.matrix (Bulinus_tot~ sTemp_Water + spH + sLat + sLon + sCond + seas_wmo+ swmo_prec + swater_speed_ms + swater_depth + sTemp_Air+ syear + duration + locality + site_irn + coll_date, data = data))= data[rows, ]
Our hypothesis is that the abundance of Bulinus species depends on the water characteristics, e.g. site_type, Temp_water, pH, Cond, swmo_prec, water_speed_ms, and water_depth. We will set the length of the collection duration as an offset.
= glm (Bulinus_tot~ offset (log (duration)) + site_type + sTemp_Water + spH + + swmo_prec + swater_speed_ms + swater_depth,data = data, family = poisson)summary (model1)
Call:
glm(formula = Bulinus_tot ~ offset(log(duration)) + site_type +
sTemp_Water + spH + sCond + swmo_prec + swater_speed_ms +
swater_depth, family = poisson, data = data)
Deviance Residuals:
Min 1Q Median 3Q Max
-11.181 -5.895 -3.382 0.729 46.751
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 0.328925 0.006417 51.258 < 2e-16 ***
site_typecanal.3 -0.161047 0.010287 -15.656 < 2e-16 ***
site_typepond -0.837273 0.022624 -37.009 < 2e-16 ***
site_typerice.p -1.378799 0.027252 -50.595 < 2e-16 ***
site_typeriver -1.730850 0.032842 -52.703 < 2e-16 ***
site_typerivulet -1.757255 0.041545 -42.298 < 2e-16 ***
site_typespillway -1.679141 0.048544 -34.590 < 2e-16 ***
sTemp_Water -0.089050 0.004435 -20.080 < 2e-16 ***
spH 0.036653 0.004501 8.144 3.82e-16 ***
sCond 0.072787 0.004979 14.620 < 2e-16 ***
swmo_prec -0.098717 0.006337 -15.577 < 2e-16 ***
swater_speed_ms -0.181606 0.007695 -23.600 < 2e-16 ***
swater_depth -0.113600 0.005998 -18.940 < 2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for poisson family taken to be 1)
Null deviance: 131410 on 2071 degrees of freedom
Residual deviance: 116126 on 2059 degrees of freedom
AIC: 122311
Number of Fisher Scoring iterations: 6
As the sites are nested within localities, we will set a nested random intercept on site_irn within locality. Also potential confounders are collection date (coll_date), the season (wet or dry months, seas_wmo), year, and maybe other environmental factors such as the air temperature?.
= glmer (Bulinus_tot~ offset (log (duration)) + site_type + sTemp_Water + spH + + swmo_prec + swater_speed_ms + swater_depth + + seas_wmo + (1 | year) + (1 | locality/ site_irn) + (swater_depth| coll_date),data = data, family = poisson)summary (model2)
Generalized linear mixed model fit by maximum likelihood (Laplace
Approximation) [glmerMod]
Family: poisson ( log )
Formula: Bulinus_tot ~ offset(log(duration)) + site_type + sTemp_Water +
spH + sCond + swmo_prec + swater_speed_ms + swater_depth +
sTemp_Air + seas_wmo + (1 | year) + (1 | locality/site_irn) +
(swater_depth | coll_date)
Data: data
AIC BIC logLik -2*log(L) df.resid
48873.9 48992.3 -24415.9 48831.9 2051
Scaled residuals:
Min 1Q Median 3Q Max
-11.997 -2.448 -0.963 1.285 41.809
Random effects:
Groups Name Variance Std.Dev. Corr
coll_date (Intercept) 1.8754 1.3694
swater_depth 1.2493 1.1177 0.29
site_irn:locality (Intercept) 0.7207 0.8489
locality (Intercept) 0.8318 0.9120
year (Intercept) 0.1613 0.4017
Number of obs: 2072, groups:
coll_date, 191; site_irn:locality, 89; locality, 20; year, 5
Fixed effects:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -0.971159 0.363661 -2.671 0.007574 **
site_typecanal.3 -0.078307 0.238497 -0.328 0.742657
site_typepond 0.291211 0.412801 0.705 0.480530
site_typerice.p -0.774537 0.343706 -2.253 0.024228 *
site_typeriver -0.532048 0.424187 -1.254 0.209741
site_typerivulet -0.448872 0.557398 -0.805 0.420647
site_typespillway -0.422082 0.560277 -0.753 0.451242
sTemp_Water -0.055643 0.018103 -3.074 0.002114 **
spH -0.029211 0.008983 -3.252 0.001147 **
sCond -0.015816 0.009465 -1.671 0.094709 .
swmo_prec -0.212222 0.099810 -2.126 0.033481 *
swater_speed_ms -0.128274 0.009414 -13.626 < 2e-16 ***
swater_depth -0.290767 0.084223 -3.452 0.000556 ***
sTemp_Air -0.102232 0.013609 -7.512 5.82e-14 ***
seas_wmowet -0.057522 0.205846 -0.279 0.779907
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Correlation matrix not shown by default, as p = 15 > 12.
Use print(x, correlation=TRUE) or
vcov(x) if you need it
Check residuals:
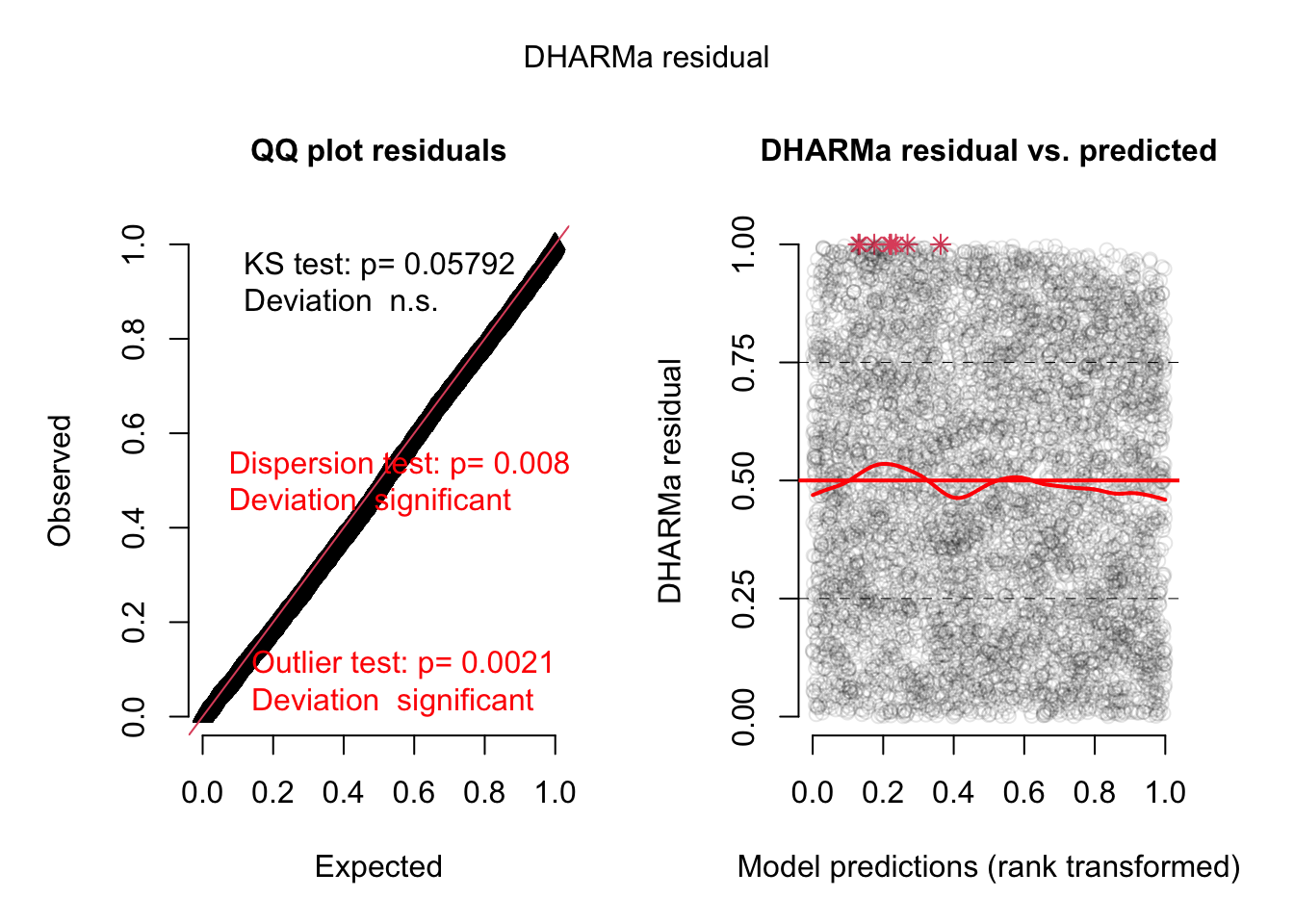
= simulateResiduals (model2, plot = TRUE , re.form = NULL )
DHARMa:testOutliers with type = binomial may have inflated Type I error rates for integer-valued distributions. To get a more exact result, it is recommended to re-run testOutliers with type = 'bootstrap'. See ?testOutliers for details
DHARMa:testOutliers with type = binomial may have inflated Type I error rates for integer-valued distributions. To get a more exact result, it is recommended to re-run testOutliers with type = 'bootstrap'. See ?testOutliers for details
Does not look great -> dispersion problems -> switch to -> negative binomial distribution:
= glmmTMB (Bulinus_tot~ offset (log (duration)) + site_type + sTemp_Water + spH + + swmo_prec + swater_speed_ms + swater_depth + + seas_wmo + (1 | year) + (1 | locality/ site_irn) + (swater_depth| coll_date),data = data, family = nbinom2)summary (model3)
Family: nbinom2 ( log )
Formula:
Bulinus_tot ~ offset(log(duration)) + site_type + sTemp_Water +
spH + sCond + swmo_prec + swater_speed_ms + swater_depth +
sTemp_Air + seas_wmo + (1 | year) + (1 | locality/site_irn) +
(swater_depth | coll_date)
Data: data
AIC BIC logLik deviance df.resid
14137.0 14261.0 -7046.5 14093.0 2050
Random effects:
Conditional model:
Groups Name Variance Std.Dev. Corr
year (Intercept) 0.32980 0.5743
site_irn:locality (Intercept) 0.59947 0.7743
locality (Intercept) 0.54314 0.7370
coll_date (Intercept) 0.66067 0.8128
swater_depth 0.02441 0.1563 0.16
Number of obs: 2072, groups:
year, 5; site_irn:locality, 89; locality, 20; coll_date, 191
Dispersion parameter for nbinom2 family (): 0.433
Conditional model:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -0.48794 0.37886 -1.288 0.197773
site_typecanal.3 -0.14448 0.24459 -0.591 0.554728
site_typepond 0.09447 0.43016 0.220 0.826172
site_typerice.p -0.92636 0.36321 -2.550 0.010757 *
site_typeriver -0.82008 0.42509 -1.929 0.053709 .
site_typerivulet -0.98675 0.54309 -1.817 0.069231 .
site_typespillway -0.41259 0.56965 -0.724 0.468888
sTemp_Water -0.10038 0.09361 -1.072 0.283532
spH -0.07251 0.05265 -1.377 0.168487
sCond 0.12188 0.05953 2.047 0.040613 *
swmo_prec -0.10279 0.07948 -1.293 0.195908
swater_speed_ms -0.12390 0.03418 -3.625 0.000289 ***
swater_depth -0.18908 0.05497 -3.440 0.000583 ***
sTemp_Air -0.01520 0.08007 -0.190 0.849444
seas_wmowet -0.03143 0.17994 -0.175 0.861355
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Check residuals:
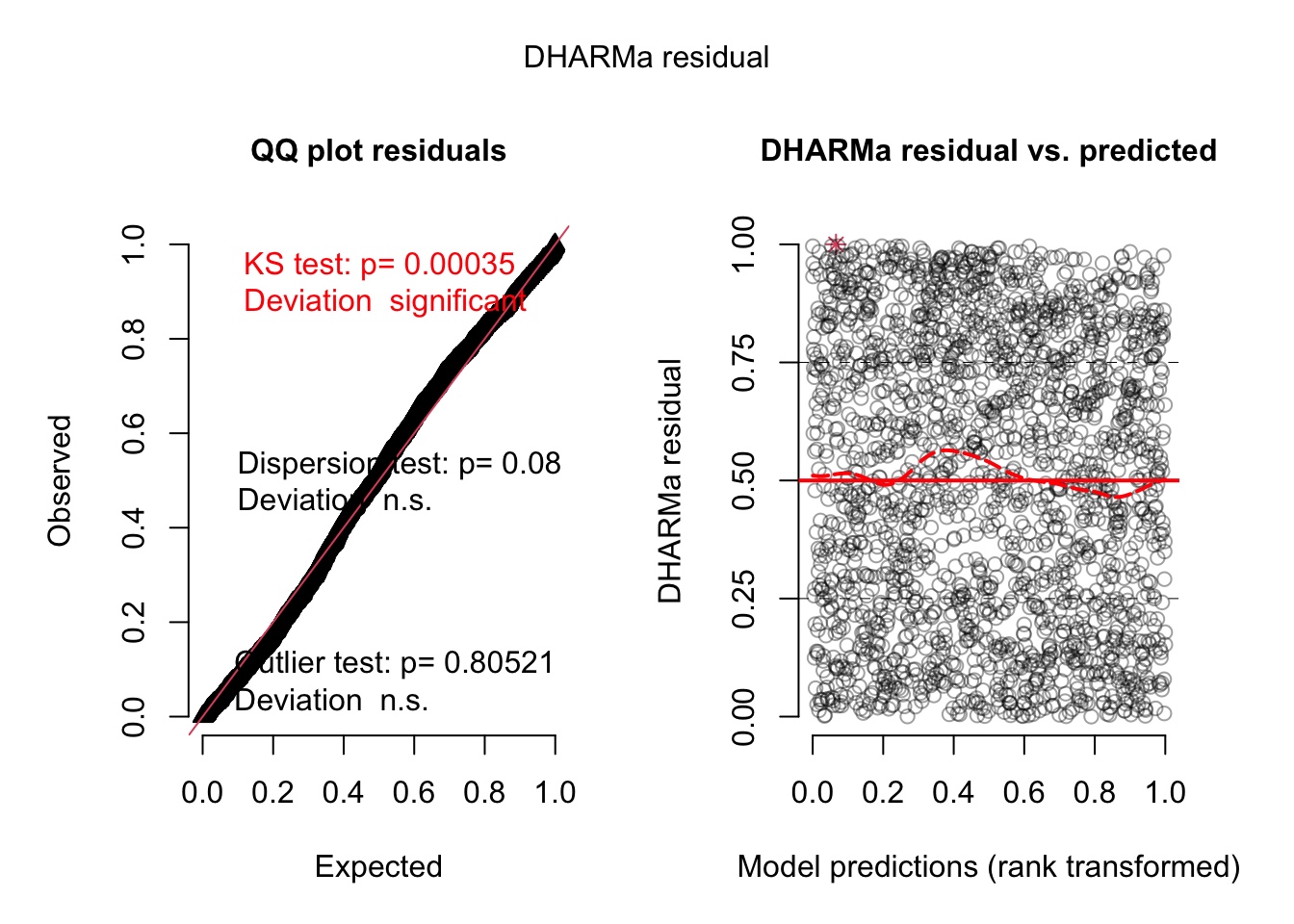
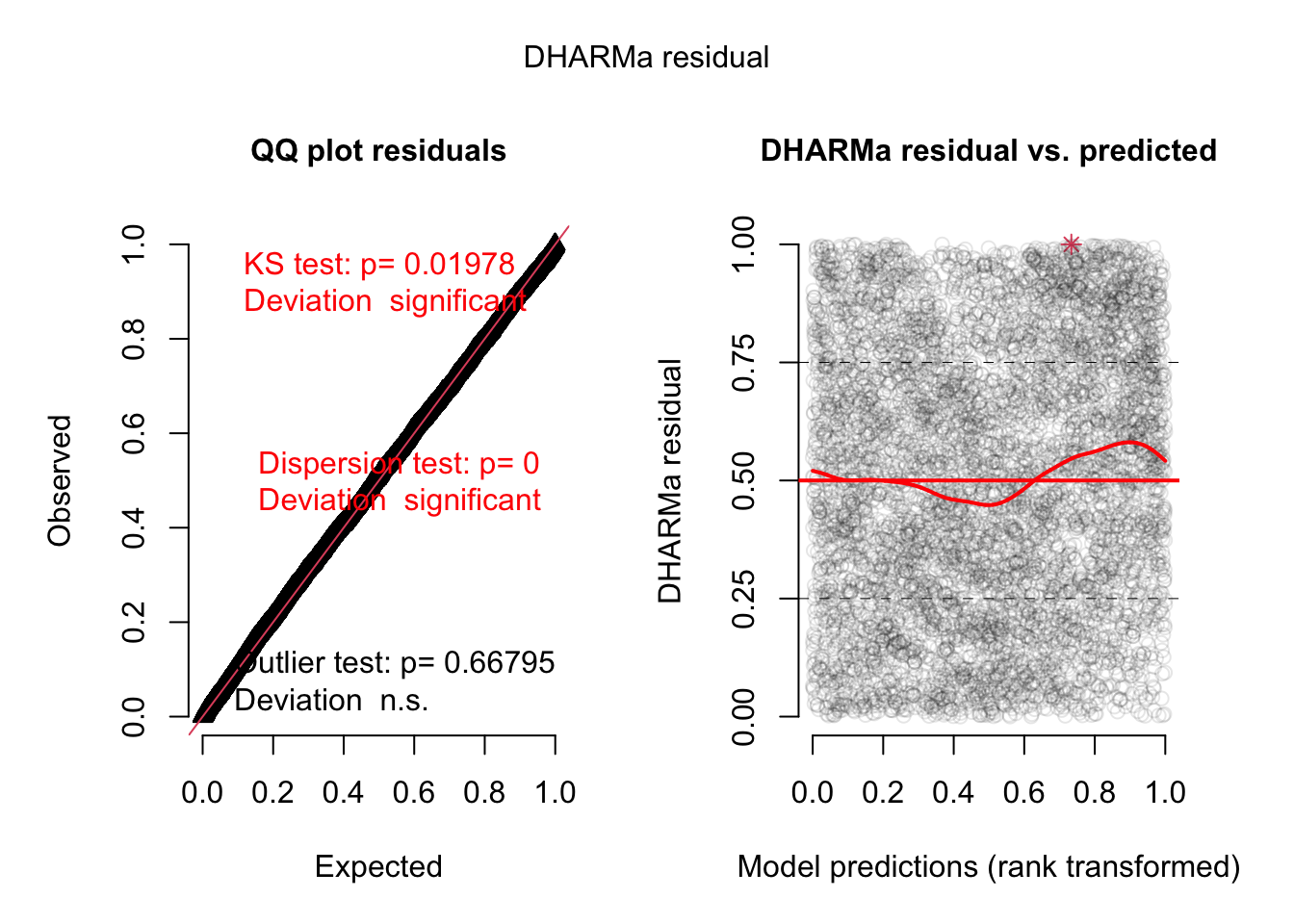
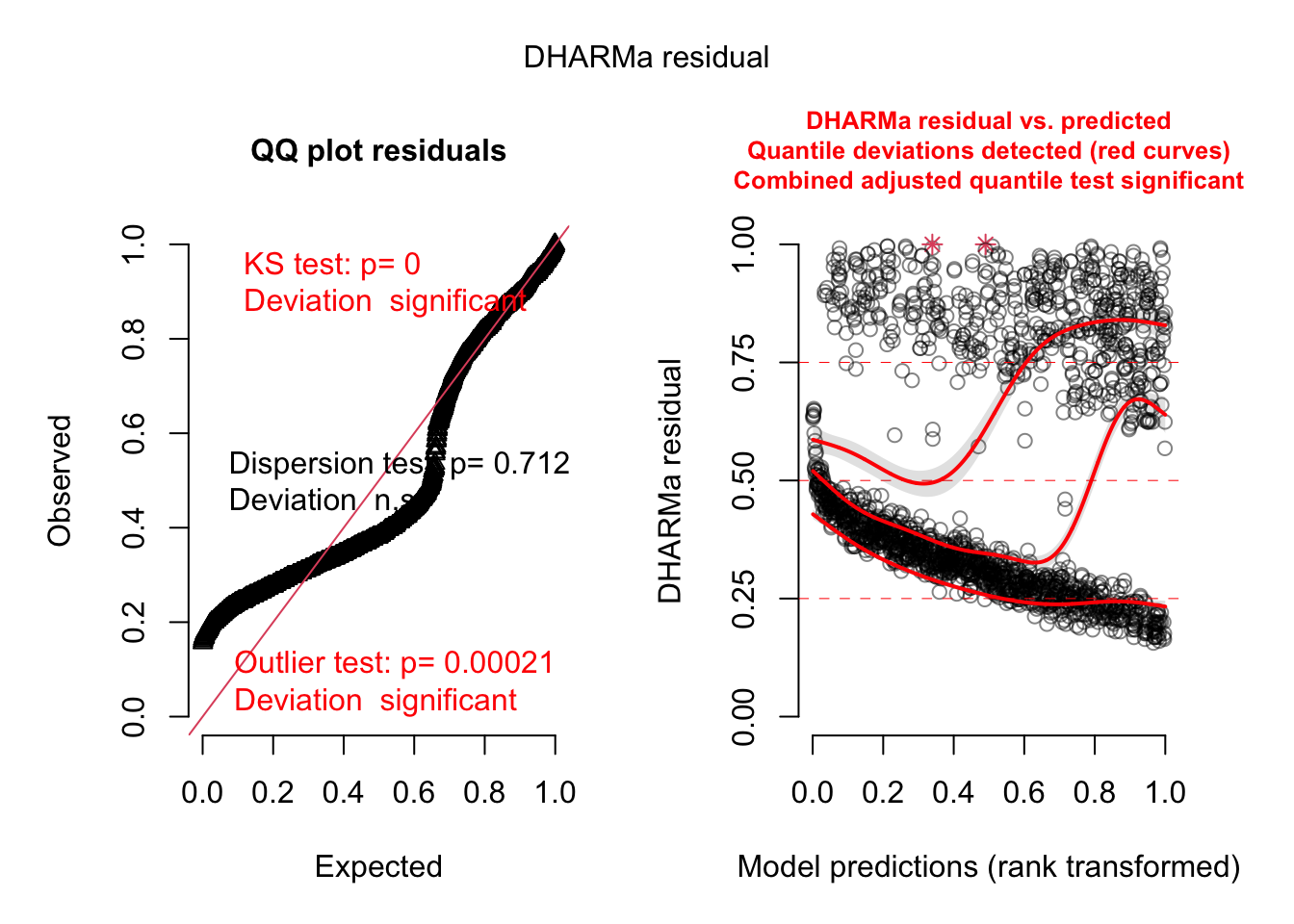
= simulateResiduals (model3, plot = TRUE )
Residuals look better but there is still a dispersion problem.
Let’s use the dispformula:
= glmmTMB (Bulinus_tot~ offset (log (duration)) + site_type + sTemp_Water + spH + + swmo_prec + swater_speed_ms + swater_depth + + seas_wmo + (1 | year) + (1 | locality/ site_irn) + (swater_depth| coll_date),dispformula = ~ swater_speed_ms + swater_depth+ sCond,data = data, family = nbinom2)summary (model4)
Family: nbinom2 ( log )
Formula:
Bulinus_tot ~ offset(log(duration)) + site_type + sTemp_Water +
spH + sCond + swmo_prec + swater_speed_ms + swater_depth +
sTemp_Air + seas_wmo + (1 | year) + (1 | locality/site_irn) +
(swater_depth | coll_date)
Dispersion: ~swater_speed_ms + swater_depth + sCond
Data: data
AIC BIC logLik deviance df.resid
14138.2 14279.1 -7044.1 14088.2 2047
Random effects:
Conditional model:
Groups Name Variance Std.Dev. Corr
year (Intercept) 0.32157 0.5671
site_irn:locality (Intercept) 0.60194 0.7758
locality (Intercept) 0.53655 0.7325
coll_date (Intercept) 0.65362 0.8085
swater_depth 0.01958 0.1399 0.14
Number of obs: 2072, groups:
year, 5; site_irn:locality, 89; locality, 20; coll_date, 191
Conditional model:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -0.46934 0.37644 -1.247 0.212485
site_typecanal.3 -0.15293 0.24497 -0.624 0.532444
site_typepond 0.05419 0.43145 0.126 0.900045
site_typerice.p -0.95968 0.36331 -2.641 0.008254 **
site_typeriver -0.84089 0.42561 -1.976 0.048184 *
site_typerivulet -1.01483 0.54260 -1.870 0.061439 .
site_typespillway -0.45053 0.57035 -0.790 0.429575
sTemp_Water -0.09780 0.09342 -1.047 0.295146
spH -0.07259 0.05246 -1.384 0.166427
sCond 0.12399 0.06344 1.954 0.050647 .
swmo_prec -0.10189 0.07895 -1.290 0.196899
swater_speed_ms -0.16899 0.04665 -3.623 0.000292 ***
swater_depth -0.17509 0.05627 -3.112 0.001861 **
sTemp_Air -0.01550 0.08014 -0.193 0.846589
seas_wmowet -0.04867 0.17952 -0.271 0.786290
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Dispersion model:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -0.838268 0.038472 -21.789 <2e-16 ***
swater_speed_ms -0.095345 0.044883 -2.124 0.0336 *
swater_depth -0.003945 0.042073 -0.094 0.9253
sCond -0.013696 0.037138 -0.369 0.7123
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
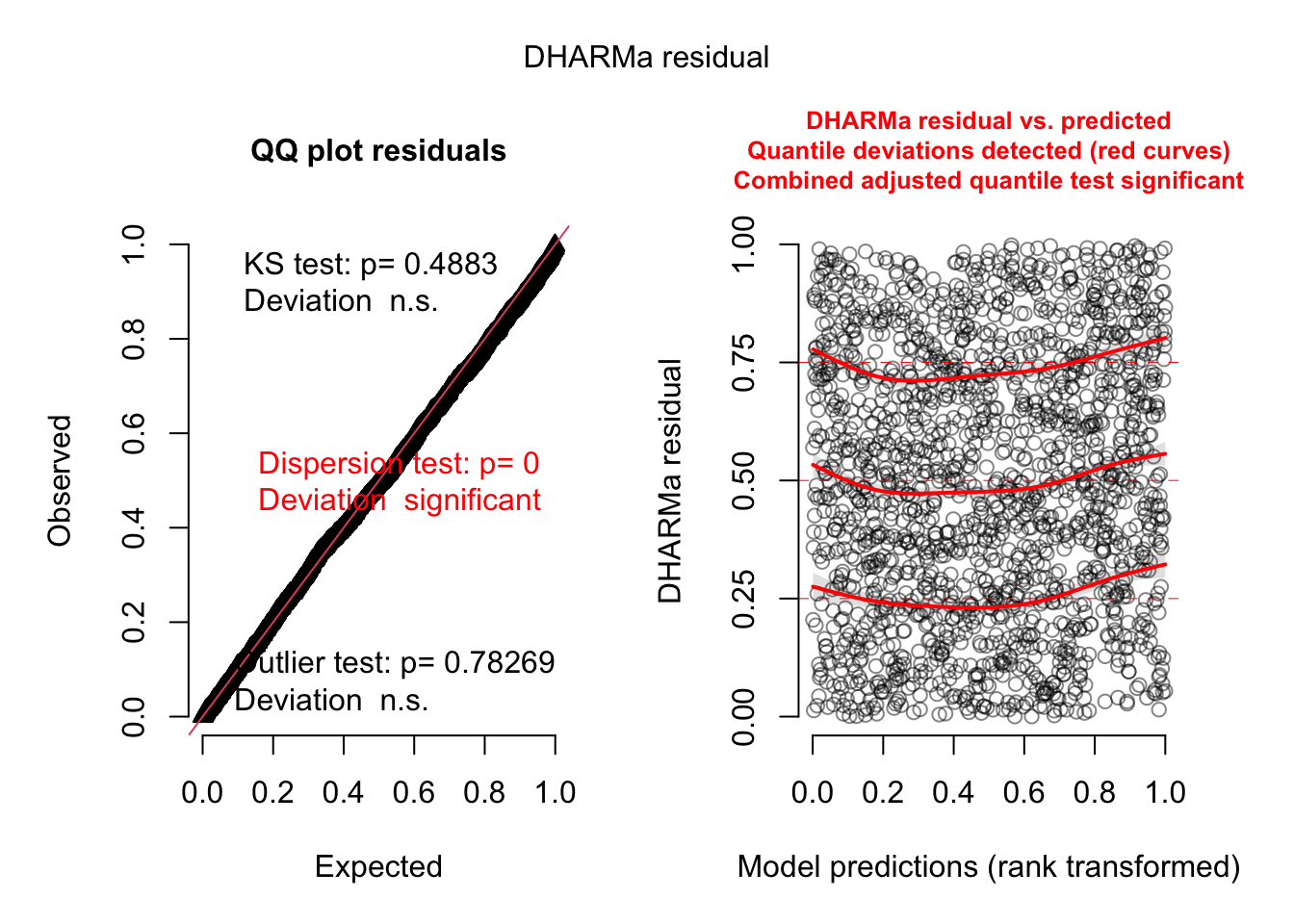
Dispformula is significant. Residuals:
= simulateResiduals (model4, plot = TRUE )
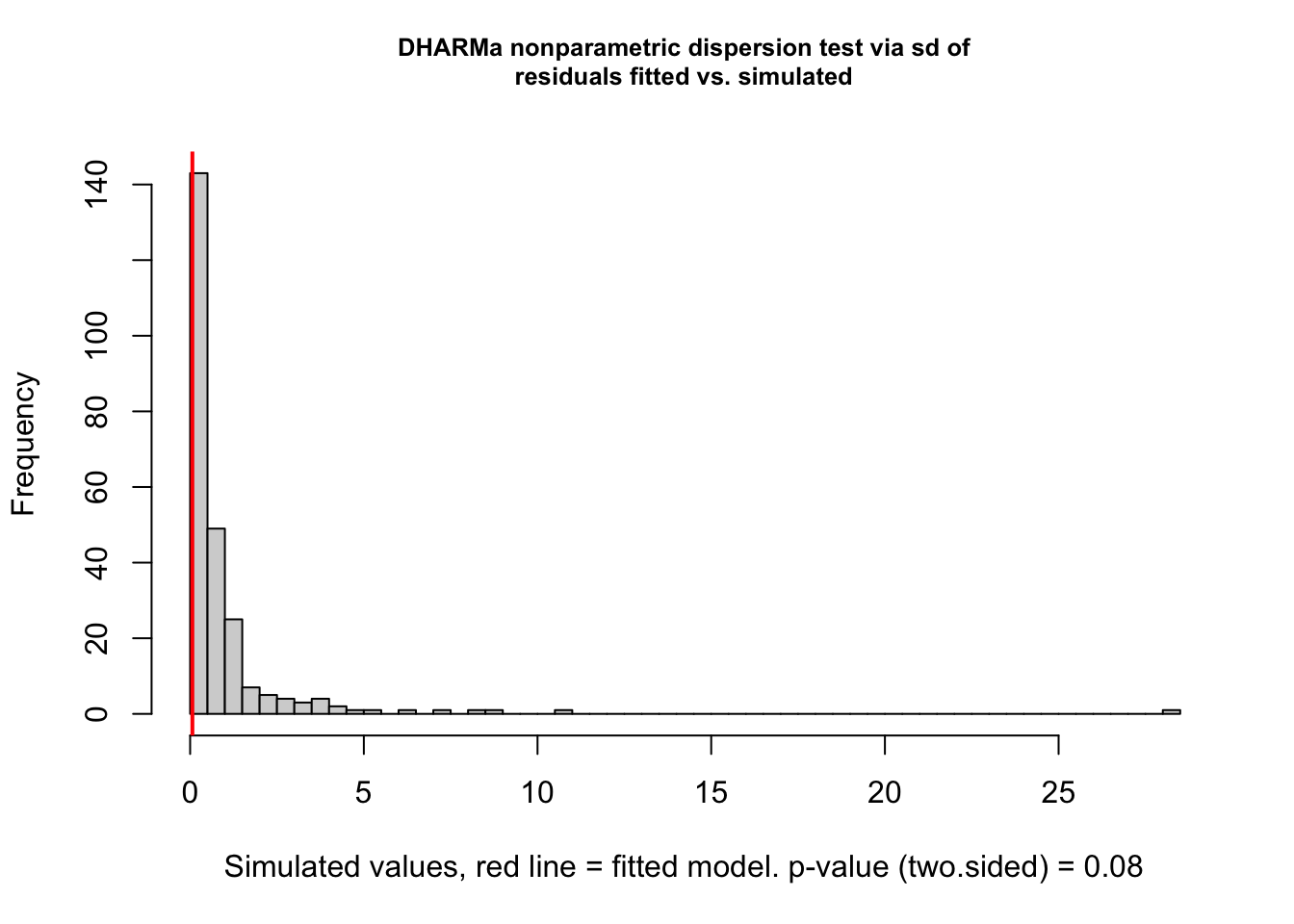
Dispersion tests are n.s.:
DHARMa nonparametric dispersion test via sd of residuals fitted vs.
simulated
data: simulationOutput
dispersion = 0.065593, p-value = 0.08
alternative hypothesis: two.sided
DHARMa zero-inflation test via comparison to expected zeros with
simulation under H0 = fitted model
data: simulationOutput
ratioObsSim = 1.052, p-value = 0.656
alternative hypothesis: two.sided
We detrended space there could be spatial autocorrelation, let’s check for it:
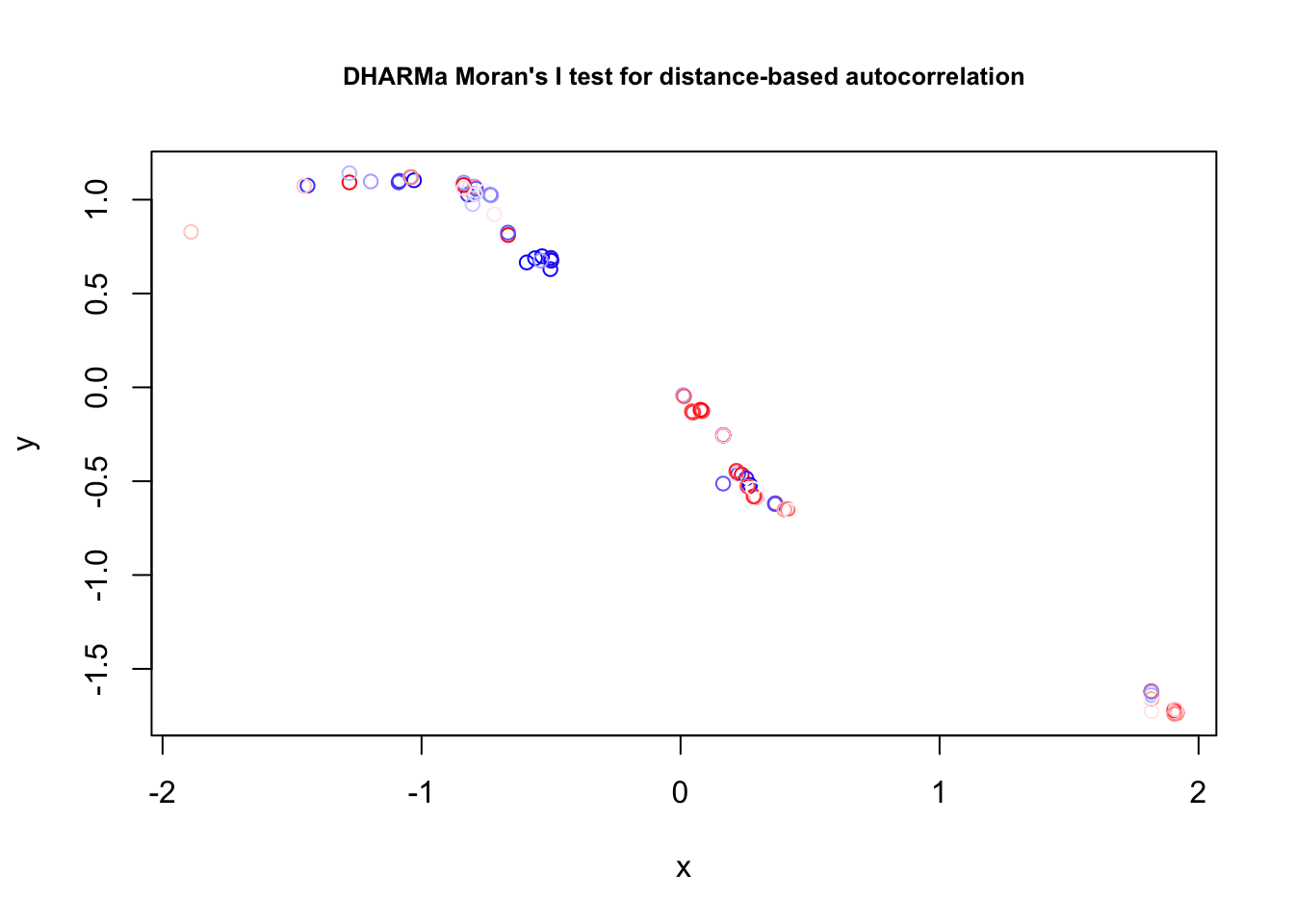
## Spatial = recalculateResiduals (res, group = c (data$ site_irn))= aggregate (cbind (data$ sLat, data$ sLon ), list ( data$ site_irn), mean)testSpatialAutocorrelation (res2, x = groupLocations$ V1, y = groupLocations$ V2)
DHARMa Moran's I test for distance-based autocorrelation
data: res2
observed = 0.295785, expected = -0.011364, sd = 0.066951, p-value =
4.482e-06
alternative hypothesis: Distance-based autocorrelation
Significant! Let’s add a spatial correlation structure:
= numFactor (data$ sLat, data$ sLon)= factor (rep (1 , nrow (data)))$ fmonth = as.factor (data$ month)= glmmTMB (Bulinus_tot~ offset (log (duration)) + site_type + sTemp_Water + spH + + swmo_prec + swater_speed_ms + swater_depth + + seas_wmo + (1 | year) + (1 | locality/ site_irn) + (swater_depth| coll_date) + exp (0 + numFac| group),dispformula = ~ swater_speed_ms + swater_depth+ sCond,data = data, family = nbinom2)
= simulateResiduals (model5, plot = TRUE )
DHARMa:testOutliers with type = binomial may have inflated Type I error rates for integer-valued distributions. To get a more exact result, it is recommended to re-run testOutliers with type = 'bootstrap'. See ?testOutliers for details
DHARMa:testOutliers with type = binomial may have inflated Type I error rates for integer-valued distributions. To get a more exact result, it is recommended to re-run testOutliers with type = 'bootstrap'. See ?testOutliers for details
glmmTMB does not support conditional simulations but we can create conditional simulations on our own:
= predict (model5, re.form = NULL , type = "response" )= predict (model5, re.form = NULL , type = "disp" )= sapply (1 : 1000 , function (i) rnbinom (length (pred),size = pred_dispersion, mu = pred))= createDHARMa (simulations, model.frame (model5)[,1 ], pred)plot (res)
Residuals do not look perfect but I would say that we can stop here now.
Prepare+scale data:
library (lme4)library (glmmTMB)library (DHARMa)= EcoData:: snails$ sTemp_Water = scale (data$ Temp_Water)$ spH = scale (data$ pH)$ swater_speed_ms = scale (data$ water_speed_ms)$ swater_depth = scale (data$ water_depth)$ sCond = scale (data$ Cond)$ swmo_prec = scale (data$ wmo_prec)$ syear = scale (data$ year)$ sLat = scale (data$ Latitude)$ sLon = scale (data$ Longitude)$ sTemp_Air = scale (data$ Temp_Air)# Let's remove NAs beforehand: = rownames (model.matrix (Bulinus_tot~ sTemp_Water + spH + sLat + sLon + sCond + seas_wmo+ swmo_prec + swater_speed_ms + swater_depth + sTemp_Air+ syear + duration + locality + site_irn + coll_date, data = data))= data[rows, ]
Let’s start directly with all potential confounders (see previous solution):
= glmmTMB (cbind (Bulinus_pos_tot, Bulinus_tot - Bulinus_pos_tot )~ offset (log (duration)) + site_type + sTemp_Water + spH + + swmo_prec + swater_speed_ms + swater_depth + + seas_wmo + (1 | year) + (1 | locality/ site_irn) + (swater_depth| coll_date),data = data, family = binomial)summary (model1)
Family: binomial ( logit )
Formula:
cbind(Bulinus_pos_tot, Bulinus_tot - Bulinus_pos_tot) ~ offset(log(duration)) +
site_type + sTemp_Water + spH + sCond + swmo_prec + swater_speed_ms +
swater_depth + sTemp_Air + seas_wmo + (1 | year) + (1 | locality/site_irn) +
(swater_depth | coll_date)
Data: data
AIC BIC logLik deviance df.resid
1323.3 1441.7 -640.7 1281.3 2051
Random effects:
Conditional model:
Groups Name Variance Std.Dev. Corr
year (Intercept) 0.04167 0.2041
site_irn:locality (Intercept) 1.79468 1.3397
locality (Intercept) 0.91215 0.9551
coll_date (Intercept) 1.40999 1.1874
swater_depth 0.46082 0.6788 0.76
Number of obs: 2072, groups:
year, 5; site_irn:locality, 89; locality, 20; coll_date, 191
Conditional model:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -9.71202 0.52321 -18.562 < 2e-16 ***
site_typecanal.3 -0.20119 0.52740 -0.381 0.70285
site_typepond 2.30894 0.81865 2.820 0.00480 **
site_typerice.p -0.21964 0.84882 -0.259 0.79582
site_typeriver -0.05417 0.99933 -0.054 0.95677
site_typerivulet 1.09537 1.05508 1.038 0.29918
site_typespillway 0.84839 1.37766 0.616 0.53802
sTemp_Water 0.02948 0.16178 0.182 0.85543
spH -0.08128 0.10741 -0.757 0.44919
sCond -0.32655 0.10527 -3.102 0.00192 **
swmo_prec -0.59112 0.32596 -1.813 0.06976 .
swater_speed_ms 0.17271 0.08010 2.156 0.03107 *
swater_depth -0.27776 0.19217 -1.445 0.14835
sTemp_Air -0.23545 0.14630 -1.609 0.10753
seas_wmowet -0.36256 0.32166 -1.127 0.25969
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
= simulateResiduals (model1, plot= TRUE )
DHARMa:testOutliers with type = binomial may have inflated Type I error rates for integer-valued distributions. To get a more exact result, it is recommended to re-run testOutliers with type = 'bootstrap'. See ?testOutliers for details
DHARMa:testOutliers with type = binomial may have inflated Type I error rates for integer-valued distributions. To get a more exact result, it is recommended to re-run testOutliers with type = 'bootstrap'. See ?testOutliers for details
We have dispersion problems, but we cannot model the dispersion for binomial models.
Check for spatial autocorrelation:
## Spatial = recalculateResiduals (res, group = c (data$ site_irn))= aggregate (cbind (data$ sLat, data$ sLon ), list ( data$ site_irn), mean)testSpatialAutocorrelation (res2, x = groupLocations$ V1, y = groupLocations$ V2)
DHARMa Moran's I test for distance-based autocorrelation
data: res2
observed = 0.159982, expected = -0.011364, sd = 0.067031, p-value =
0.01058
alternative hypothesis: Distance-based autocorrelation
Significant! Let’s correct for spatial autocorrelation with a correlation structure:
= numFactor (data$ sLat, data$ sLon)= factor (rep (1 , nrow (data)))$ fmonth = as.factor (data$ month)= glmmTMB (cbind (Bulinus_pos_tot, Bulinus_tot - Bulinus_pos_tot )~ offset (log (duration)) + site_type + sTemp_Water + spH + + swmo_prec + swater_speed_ms + swater_depth + + seas_wmo + (1 | year) + (1 | locality/ site_irn) + (swater_depth| coll_date) + exp (0 + numFac| group),data = data, family = binomial)
= simulateResiduals (model2, plot = TRUE )
They look good now!
The species models are connected by their response to latent variable (unobserved environment). For that, we will transform our dataset with respect to species from wide (sp1, sp2, sp3) to long format (species abundances in one column and a second column telling us the group (species)). In the model then, we will separate the species and their responses by using ~0+Species + Species:(predictors).
The latent variable structure is set by the rr(…) object in the formula:
library (lme4)library (glmmTMB)library (DHARMa)library (tidyverse)= EcoData:: snails$ sTemp_Water = scale (data$ Temp_Water)$ spH = scale (data$ pH)$ swater_speed_ms = scale (data$ water_speed_ms)$ swater_depth = scale (data$ water_depth)$ sCond = scale (data$ Cond)$ swmo_prec = scale (data$ wmo_prec)$ syear = scale (data$ year)$ sLat = scale (data$ Latitude)$ sLon = scale (data$ Longitude)$ sTemp_Air = scale (data$ Temp_Air)# Let's remove NAs beforehand: = rownames (model.matrix (cbind (Bulinus_pos_tot, Bulinus_tot- Bulinus_pos_tot)~ sTemp_Water + spH + sLat + sLon + sCond + seas_wmo+ swmo_prec + swater_speed_ms + swater_depth + sTemp_Air+ syear + duration + locality + site_irn + coll_date, data = data))= data[rows, ]= %>% pivot_longer (cols = c ("BP_tot" , "BF_tot" , "BT_tot" ),names_to = "Species" ,values_to = "Abundance" )= numFactor (data$ sLat, data$ sLon)= factor (rep (1 , nrow (data)))= numFactor (data$ sLat, data$ sLon)= factor (rep (1 , nrow (data)))$ fmonth = as.factor (data$ month)= glmmTMB (Abundance~ 0 + offset (log (duration)) + Species + Species: (site_type + + spH + sCond + swmo_prec + swater_speed_ms + swater_depth + + seas_wmo) + (1 | year) + (1 | locality/ site_irn) + (swater_depth| coll_date: Species) + exp (0 + numFac| group) + rr (Species + 0 | locality: site_irn, d = 2 ),dispformula = ~ 0 + Species+ Species: (swater_speed_ms + swater_depth+ sCond),data = data, family = nbinom2)
Warning in finalizeTMB(TMBStruc, obj, fit, h, data.tmb.old): Model convergence
problem; singular convergence (7). See vignette('troubleshooting'),
help('diagnose')
Unconditional residuals:
plot (simulateResiduals (modelJoint))
Conditional residuals:
= predict (modelJoint, re.form = NULL , type = "response" )= predict (modelJoint, re.form = NULL , type = "disp" )= sapply (1 : 1000 , function (i) rnbinom (length (pred),size = pred_dispersion, mu = pred))= createDHARMa (simulations, model.frame (modelJoint)[,1 ], pred)plot (res)
Seed bank
library (EcoData)library (glmmTMB)library (lme4)library (lmerTest)library (DHARMa)library (tidyverse):: seedBank
The seedBank data set in the EcoData package includes observation on seed bank presence and size in different vegetaition plots.
The scientific question is if the ability of plants to build a seed bank depends on their seed traits (see help).
The data also contains data on environmental factors and plant traits. You should consider if it is useful to add them to the analysis.
Tasks:
Fit a lm/lmm with SBDensity as response
Bonus: Add phylogenetic correlation structure
Fit a glm/glmm with SBPA as response (Bonus: add phylogenetic correlation structure)
Prepare+scale data:
= as.data.frame (EcoData:: seedBank)$ sAltitude = scale (data$ Altitude)$ sSeedMass = scale (data$ SeedMass)$ sSeedShape = scale (data$ SeedShape)$ sSeedN = scale (data$ SeedN)$ sSeedPr = scale (data$ SeedPr)$ sDormRank = scale (data$ DormRank)$ sTemp = scale (data$ Temp)$ sHum = scale (data$ Humidity)$ sNitro = scale (data$ Nitrogen)$ sGrazing = scale (data$ Grazing)$ sMGT = scale (data$ MGT)$ sJwidth = scale (data$ Jwidth)$ sEpiStein = scale (data$ EpiStein)$ sMGR = scale (data$ MGR)$ sT95 = scale (data$ T95)# Let's remove NAs beforehand: = rownames (model.matrix (SBDensity~ sAltitude + sSeedMass + sSeedShape + sSeedN + + sDormRank + sTemp + sHum + sNitro + sMGT + + sEpiStein + sT95 + + sGrazing + Site + Species, data = data))= data[rows, ]
The response is highly skewed, so it makes sense to log-transform:
$ logSBDensity = log (data$ SBDensity + 1 )
Let’s fit a base model with with our hypothesis that logSBDensity ~ sSeedMass (Seed Mass) + sSeedShape + sSeedN (Number of Seeds).
We set random intercepts on species and sites because we assume that there are species and site specific variations:
= lmer (logSBDensity~ + sSeedShape + sSeedN + 1 | Site) + (1 | Species),data = data)summary (model1)
Linear mixed model fit by REML. t-tests use Satterthwaite's method [
lmerModLmerTest]
Formula: logSBDensity ~ sSeedMass + sSeedShape + sSeedN + (1 | Site) +
(1 | Species)
Data: data
REML criterion at convergence: 7531.9
Scaled residuals:
Min 1Q Median 3Q Max
-3.6261 -0.4945 -0.0823 0.4542 3.1525
Random effects:
Groups Name Variance Std.Dev.
Species (Intercept) 3.0724 1.7528
Site (Intercept) 0.4705 0.6859
Residual 3.6790 1.9181
Number of obs: 1729, groups: Species, 152; Site, 17
Fixed effects:
Estimate Std. Error df t value Pr(>|t|)
(Intercept) 1.5314 0.2271 42.4393 6.743 3.23e-08 ***
sSeedMass -0.3316 0.1273 175.6509 -2.604 0.00999 **
sSeedShape -0.3175 0.1546 155.2402 -2.054 0.04168 *
sSeedN -0.1818 0.1289 165.3141 -1.410 0.16028
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Correlation of Fixed Effects:
(Intr) sSdMss sSdShp
sSeedMass -0.037
sSeedShape -0.034 0.168
sSeedN -0.031 0.039 0.077
Environmental factors can be potential confounders, let’s add them to the model:
= lmer (logSBDensity~ + sSeedShape + sSeedN + + sHum + 1 | Site) + (1 | Species),data = data)summary (model2)
Linear mixed model fit by REML. t-tests use Satterthwaite's method [
lmerModLmerTest]
Formula: logSBDensity ~ sSeedMass + sSeedShape + sSeedN + sAltitude +
sHum + (1 | Site) + (1 | Species)
Data: data
REML criterion at convergence: 7510.7
Scaled residuals:
Min 1Q Median 3Q Max
-3.6468 -0.5089 -0.0720 0.4388 3.2140
Random effects:
Groups Name Variance Std.Dev.
Species (Intercept) 3.03405 1.7419
Site (Intercept) 0.06929 0.2632
Residual 3.68099 1.9186
Number of obs: 1729, groups: Species, 152; Site, 17
Fixed effects:
Estimate Std. Error df t value Pr(>|t|)
(Intercept) 1.65385 0.16664 113.38609 9.925 < 2e-16 ***
sSeedMass -0.32886 0.12662 176.33088 -2.597 0.0102 *
sSeedShape -0.32165 0.15372 155.68190 -2.092 0.0380 *
sSeedN -0.18051 0.12816 165.92065 -1.408 0.1609
sAltitude -0.56452 0.10125 17.10538 -5.575 3.27e-05 ***
sHum 0.14280 0.09738 14.04198 1.466 0.1646
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Correlation of Fixed Effects:
(Intr) sSdMss sSdShp sSeedN sAlttd
sSeedMass -0.052
sSeedShape -0.044 0.168
sSeedN -0.042 0.039 0.077
sAltitude -0.058 0.007 -0.012 -0.006
sHum -0.004 -0.008 -0.003 0.004 0.512
sSeedShape and sSeedN are now statistically significant!
Question :
What about the germination temperatures, T50/T95?
Answer:
They could be mediators, Seed Shape -> T95 -> SBDensity, so it is up to if you want to include them or not!
Residual checks:
Check for missing random slopes:
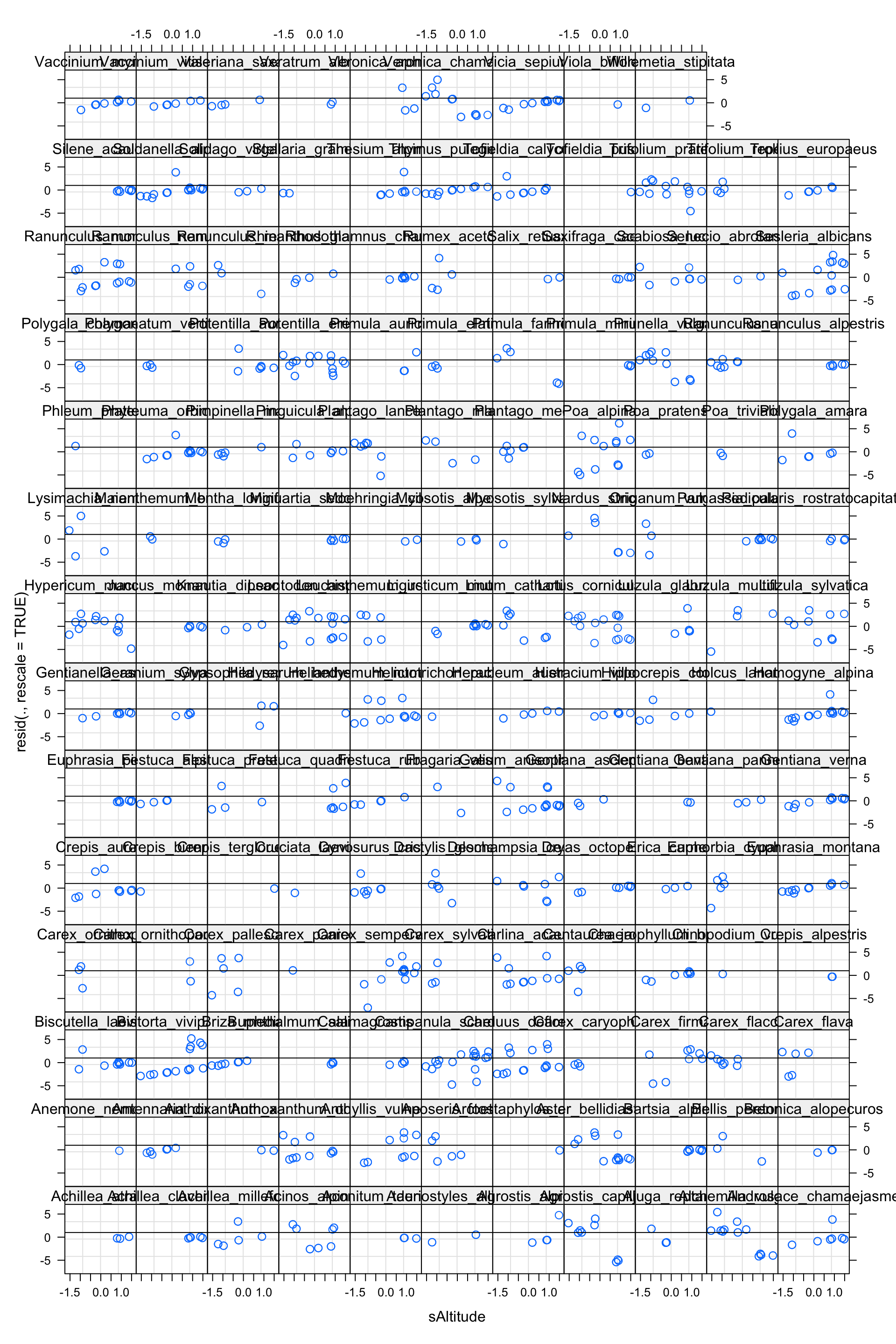
plot (model2, resid (., rescale= TRUE ) ~ fitted (.) | Species, abline = 1 )
There seems to be a pattern!
plot (model2, resid (., rescale= TRUE ) ~ sAltitude | Species, abline = 1 )
The pattern seems to be caused by sAltitude (check plot(model2, resid(., rescale=TRUE) ~ sSeedMass | Species, abline = 1) )
Random slope model
Let’s add a random slope on sAlitude:
= lmer (logSBDensity~ + sSeedShape + sSeedN + + sHum + 1 | Site) + (sAltitude| Species),data = data)summary (model3)
Linear mixed model fit by REML. t-tests use Satterthwaite's method [
lmerModLmerTest]
Formula: logSBDensity ~ sSeedMass + sSeedShape + sSeedN + sAltitude +
sHum + (1 | Site) + (sAltitude | Species)
Data: data
REML criterion at convergence: 7216.1
Scaled residuals:
Min 1Q Median 3Q Max
-2.77162 -0.39884 -0.08256 0.22365 3.06745
Random effects:
Groups Name Variance Std.Dev. Corr
Species (Intercept) 2.61864 1.6182
sAltitude 1.31560 1.1470 -0.42
Site (Intercept) 0.09023 0.3004
Residual 2.79664 1.6723
Number of obs: 1729, groups: Species, 152; Site, 17
Fixed effects:
Estimate Std. Error df t value Pr(>|t|)
(Intercept) 1.52436 0.16898 106.79022 9.021 8.45e-15 ***
sSeedMass -0.29923 0.13843 181.27154 -2.162 0.031960 *
sSeedShape -0.25730 0.14094 145.11888 -1.826 0.069969 .
sSeedN -0.15018 0.11145 147.11514 -1.347 0.179918
sAltitude -0.59612 0.15103 54.56729 -3.947 0.000228 ***
sHum 0.09296 0.10330 13.65567 0.900 0.383740
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Correlation of Fixed Effects:
(Intr) sSdMss sSdShp sSeedN sAlttd
sSeedMass -0.042
sSeedShape -0.052 0.139
sSeedN -0.045 0.037 0.086
sAltitude -0.286 0.040 -0.021 -0.003
sHum 0.015 -0.009 -0.009 0.004 0.367
Residual checks:
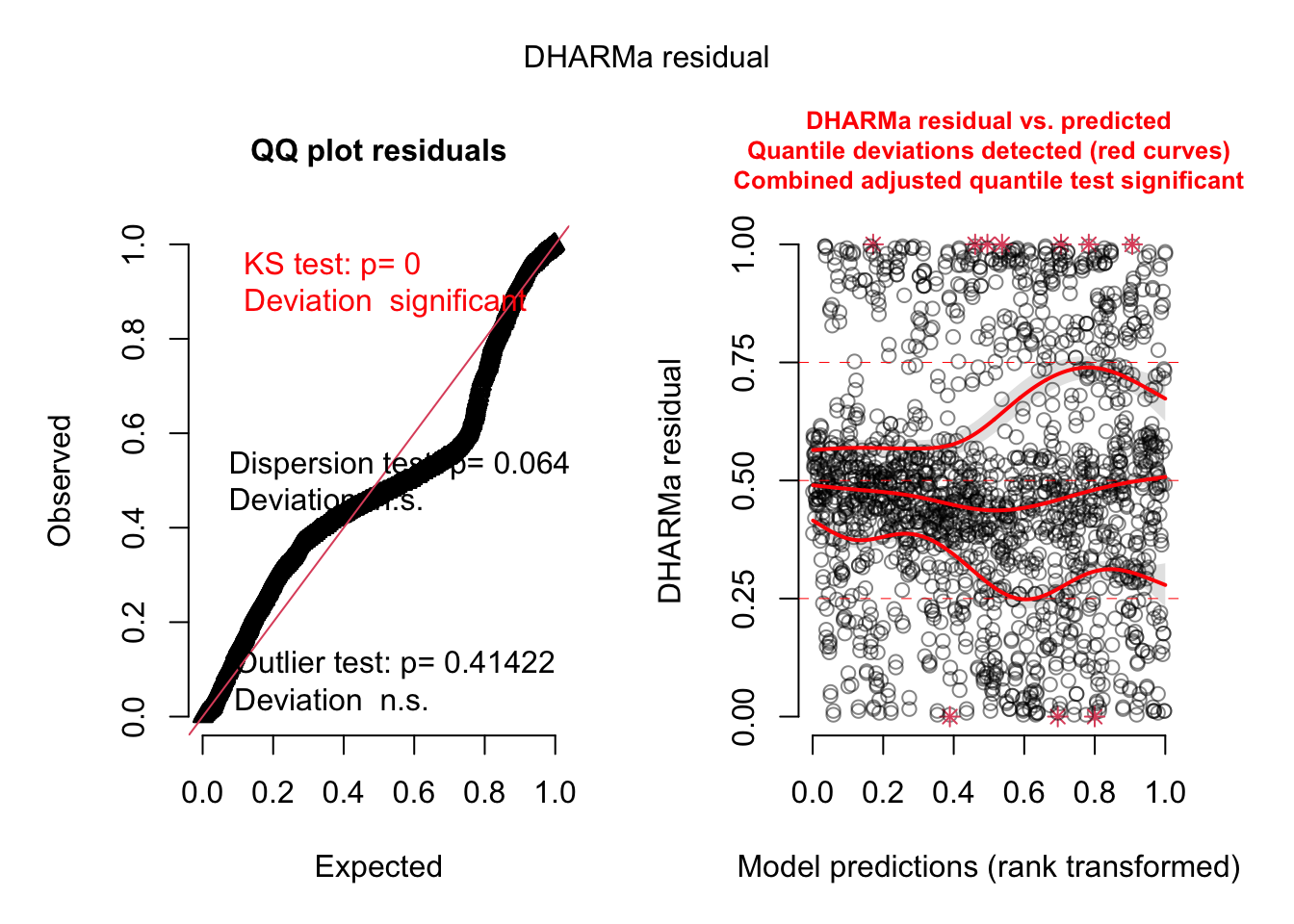
plot (simulateResiduals (model3, re.form= NULL ))
There seems to be dispersion problem.
Bonus: Modeling variance with glmmTMB
= glmmTMB (logSBDensity~ + sSeedShape + sSeedN + + sHum + 1 | Site) + (sAltitude| Species),dispformula = ~ sAltitude + sHum,data = data)summary (model4)
Family: gaussian ( identity )
Formula: logSBDensity ~ sSeedMass + sSeedShape + sSeedN + sAltitude +
sHum + (1 | Site) + (sAltitude | Species)
Dispersion: ~sAltitude + sHum
Data: data
AIC BIC logLik deviance df.resid
7196.3 7267.2 -3585.2 7170.3 1716
Random effects:
Conditional model:
Groups Name Variance Std.Dev. Corr
Site (Intercept) 0.07148 0.2674
Species (Intercept) 2.53442 1.5920
sAltitude 1.30195 1.1410 -0.44
Residual NA NA
Number of obs: 1729, groups: Site, 17; Species, 152
Conditional model:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 1.51412 0.16438 9.211 < 2e-16 ***
sSeedMass -0.28914 0.13762 -2.101 0.0356 *
sSeedShape -0.24086 0.13832 -1.741 0.0816 .
sSeedN -0.14539 0.10827 -1.343 0.1793
sAltitude -0.59734 0.14562 -4.102 4.09e-05 ***
sHum 0.09070 0.09475 0.957 0.3384
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Dispersion model:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 0.51449 0.01856 27.713 < 2e-16 ***
sAltitude -0.12343 0.02397 -5.148 2.63e-07 ***
sHum -0.01575 0.02630 -0.599 0.549
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Prepare phylogeny:
The following object is masked from 'package:dplyr':
where
Attaching package: 'geiger'
The following object is masked from 'package:brms':
bf
= unique (data$ Species)= data.frame (Species = species)rownames (species_df) = species= name.check (plantPhylo, species_df)# drop rest of the species = drop.tip (plantPhylo, obj$ tree_not_data)summary (phyl.upd)
Phylogenetic tree: phyl.upd
Number of tips: 152
Number of nodes: 140
Branch lengths:
mean: 22.21203
variance: 624.9334
distribution summary:
Min. 1st Qu. Median 3rd Qu. Max.
0.200000 5.425002 12.800003 30.402499 123.000001
Root edge: 1
First ten tip labels: Tofieldia_pusilla
Tofieldia_calyculata
Veratrum_album
Maianthemum_bifolium
Polygonatum_verticillatum
Juncus_monanthos
Luzula_glabrata
Luzula_sylvatica
Luzula_multiflora
Carex_sempervirens
First ten node labels: N398
N401
Tofieldiaceae
N573
N636
N1019
N1054
N1063
Juncaceae
Luzula
# check the names in the tree and in the data set name.check (phyl.upd, species_df)= multi2di (phyl.upd)
nlme:
library (nlme)= gls (logSBDensity ~ + sSeedShape + sSeedN + + sHum,correlation = corBrownian (phy = phyl.upd2, form = ~ Species),data = data)summary (model4)
Generalized least squares fit by REML
Model: logSBDensity ~ sSeedMass + sSeedShape + sSeedN + sAltitude + sHum
Data: data
AIC BIC logLik
-300576.2 -300538 150295.1
Correlation Structure: corBrownian
Formula: ~Species
Parameter estimate(s):
numeric(0)
Coefficients:
Value Std.Error t-value p-value
(Intercept) 1.8940733 0.7879503 2.403798 0.0163
sSeedMass -0.3233717 0.0930947 -3.473578 0.0005
sSeedShape -0.1898476 0.1184347 -1.602973 0.1091
sSeedN -0.0678053 0.1180418 -0.574418 0.5658
sAltitude -0.6393685 0.1998323 -3.199526 0.0014
sHum 0.0454568 0.1819503 0.249831 0.8027
Correlation:
(Intr) sSdMss sSdShp sSeedN sAlttd
sSeedMass -0.117
sSeedShape 0.041 0.048
sSeedN -0.007 0.017 0.037
sAltitude 0.167 -0.012 -0.008 0.007
sHum 0.083 0.013 0.000 -0.007 0.621
Standardized residuals:
Min Q1 Med Q3 Max
-1.4951026 -0.8482715 -0.5644143 1.1540499 4.1762125
Residual standard error: 2.144554
Degrees of freedom: 1729 total; 1723 residual
glmmTMB (not perfect because lack of support of specific phylogenetic correlation structures):
= ape:: cophenetic.phylo (phyl.upd2) # create distance matrix = vcv (phyl.upd2)[unique (data$ Species), unique (data$ Species)]### #the following code was taken from https://github.com/glmmTMB/glmmTMB/blob/master/misc/fixcorr.rmd <- function (Sigma,corrs.only= FALSE ) {<- log (diag (Sigma))/ 2 <- cov2cor (Sigma)<- chol (cr)<- cc %*% diag (1 / diag (cc))<- cc[upper.tri (cc)]if (corrs.only) return (corrs)<- c (logsd,corrs)return (ret)= as.theta.vcov (correlation_matrix, corrs.only= TRUE )##### $ dummy = factor (rep (0 , nrow (data)))= length (unique (data$ Species))= glmmTMB (logSBDensity~ + sSeedShape + sSeedN + + sHum + 1 | Site) + (sAltitude| Species) + 1 + Species| dummy),dispformula = ~ sAltitude + sHum,map= list (theta= factor (c (rep (0 , 4 ), rep (1 ,nsp),rep (NA ,length (corrs))) )),start= list (theta= c (rep (0 , 4 ), rep (0 ,nsp),corrs)),data = data)summary (model5)
Family: gaussian ( identity )
Formula: logSBDensity ~ sSeedMass + sSeedShape + sSeedN + sAltitude +
sHum + (1 | Site) + (sAltitude | Species) + (1 + Species | dummy)
Dispersion: ~sAltitude + sHum
Data: data
AIC BIC logLik deviance df.resid
7264.6 7324.6 -3621.3 7242.6 1718
Random effects:
Conditional model:
Groups Name Variance Std.Dev. Corr
Site (Intercept) 1.022 1.011
Species (Intercept) 1.022 1.011
sAltitude 1.022 1.011 0.01
dummy (Intercept) 2.394 1.547
SpeciesAchillea_clavennae 2.394 1.547 0.00
SpeciesAchillea_millefolium 2.394 1.547 0.20 0.00
SpeciesAcinos_alpinus 2.394 1.547 0.28 0.00 0.20
SpeciesAconitum_tauricum 2.394 1.547 0.31 0.00 0.20
SpeciesAdenostyles_alliariae 2.394 1.547 0.00 0.93 0.00
SpeciesAgrostis_alpina 2.394 1.547 0.20 0.00 0.34
SpeciesAgrostis_capillaris 2.394 1.547 0.31 0.00 0.20
SpeciesAjuga_reptans 2.394 1.547 0.31 0.00 0.20
SpeciesAlchemilla_vulgaris 2.394 1.547 0.20 0.00 0.26
SpeciesAndrosace_chamaejasme 2.394 1.547 0.25 0.00 0.20
SpeciesAnemone_nemorosa 2.394 1.547 0.00 0.93 0.00
SpeciesAntennaria_dioica 2.394 1.547 0.31 0.00 0.20
SpeciesAnthoxanthum_alpinum 2.394 1.547 0.31 0.00 0.20
SpeciesAnthoxanthum_odoratum 2.394 1.547 0.00 0.45 0.00
SpeciesAnthyllis_vulneraria 2.394 1.547 0.00 0.45 0.00
SpeciesAposeris_foetida 2.394 1.547 0.00 0.45 0.00
SpeciesArctostaphylos_alpinus 2.394 1.547 0.00 0.45 0.00
SpeciesAster_bellidiastrum 2.394 1.547 0.00 0.45 0.00
SpeciesBartsia_alpina 2.394 1.547 0.00 0.45 0.00
SpeciesBellis_perennis 2.394 1.547 0.00 0.45 0.00
SpeciesBetonica_alopecuros 2.394 1.547 0.00 0.45 0.00
SpeciesBiscutella_laevigata 2.394 1.547 0.00 0.45 0.00
SpeciesBistorta_vivipara 2.394 1.547 0.31 0.00 0.20
SpeciesBriza_media 2.394 1.547 0.31 0.00 0.20
SpeciesBuphthalmum_salicifolium 2.394 1.547 0.31 0.00 0.20
SpeciesCalamagrostis_varia 2.394 1.547 0.00 0.88 0.00
SpeciesCampanula_scheuchzeri 2.394 1.547 0.00 0.88 0.00
SpeciesCarduus_defloratus 2.394 1.547 0.20 0.00 0.51
SpeciesCarex_caryophyllea 2.394 1.547 0.20 0.00 0.30
SpeciesCarex_firma 2.394 1.547 0.55 0.00 0.20
SpeciesCarex_flacca 2.394 1.547 0.00 0.88 0.00
SpeciesCarex_flava 2.394 1.547 0.40 0.00 0.20
SpeciesCarex_ornithopoda 2.394 1.547 0.40 0.00 0.20
SpeciesCarex_ornithopodioides 2.394 1.547 0.20 0.00 0.26
SpeciesCarex_pallescens 2.394 1.547 0.20 0.00 0.34
SpeciesCarex_panicea 2.394 1.547 0.31 0.00 0.20
SpeciesCarex_sempervirens 2.394 1.547 0.20 0.00 0.30
SpeciesCarex_sylvatica 2.394 1.547 0.31 0.00 0.20
SpeciesCarlina_acaulis 2.394 1.547 0.31 0.00 0.20
SpeciesCentaurea_jacea 2.394 1.547 0.20 0.00 0.30
SpeciesChaerophyllum_hirsutum 2.394 1.547 0.20 0.00 0.34
SpeciesClinopodium_vulgare 2.394 1.547 0.00 0.17 0.00
SpeciesCrepis_alpestris 2.394 1.547 0.85 0.00 0.20
SpeciesCrepis_aurea 2.394 1.547 0.31 0.00 0.20
SpeciesCrepis_biennis 2.394 1.547 0.55 0.00 0.20
SpeciesCrepis_terglouensis 2.394 1.547 0.55 0.00 0.20
SpeciesCruciata_laevipes 2.394 1.547 0.55 0.00 0.20
SpeciesCynosurus_cristatus 2.394 1.547 0.55 0.00 0.20
SpeciesDactylis_glomerata 2.394 1.547 0.00 0.88 0.00
SpeciesDeschampsia_cespitosa 2.394 1.547 0.00 0.88 0.00
SpeciesDryas_octopetala 2.394 1.547 0.00 0.88 0.00
SpeciesErica_carnea 2.394 1.547 0.20 0.00 0.34
SpeciesEuphorbia_cyparissias 2.394 1.547 0.20 0.00 0.34
SpeciesEuphrasia_montana 2.394 1.547 0.00 0.17 0.00
SpeciesEuphrasia_picta 2.394 1.547 0.20 0.00 0.78
SpeciesFestuca_alpina 2.394 1.547 0.28 0.00 0.20
SpeciesFestuca_pratensis 2.394 1.547 0.28 0.00 0.20
SpeciesFestuca_quadriflora 2.394 1.547 0.69 0.00 0.20
SpeciesFestuca_rubra 2.394 1.547 0.09 0.00 0.09
SpeciesFragaria_vesca 2.394 1.547 0.09 0.00 0.09
SpeciesGalium_anisophyllon 2.394 1.547 0.09 0.00 0.09
SpeciesGentiana_asclepiadea 2.394 1.547 0.31 0.00 0.20
SpeciesGentiana_bavarica 2.394 1.547 0.00 0.88 0.00
SpeciesGentiana_pannonica 2.394 1.547 0.00 0.11 0.00
SpeciesGentiana_verna 2.394 1.547 0.20 0.00 0.34
SpeciesGentianella_aspera 2.394 1.547 0.20 0.00 0.34
SpeciesGeranium_sylvaticum 2.394 1.547 0.31 0.00 0.20
SpeciesGypsophila_repens 2.394 1.547 0.55 0.00 0.20
SpeciesHedysarum_hedysaroides 2.394 1.547 0.55 0.00 0.20
SpeciesHelianthemum_nummularium 2.394 1.547 0.31 0.00 0.20
SpeciesHelictotrichon_pubescens 2.394 1.547 0.00 0.94 0.00
SpeciesHeracleum_austriacum 2.394 1.547 0.00 0.45 0.00
SpeciesHieracium_villosum 2.394 1.547 0.31 0.00 0.20
SpeciesHippocrepis_comosa 2.394 1.547 0.00 0.88 0.00
SpeciesHolcus_lanatus 2.394 1.547 0.40 0.00 0.20
SpeciesHomogyne_alpina 2.394 1.547 0.20 0.00 0.26
SpeciesHypericum_maculatum 2.394 1.547 0.00 0.45 0.00
SpeciesJuncus_monanthos 2.394 1.547 0.00 0.45 0.00
SpeciesKnautia_dipsacifolia 2.394 1.547 0.00 0.45 0.00
SpeciesLeontodon_hispidus 2.394 1.547 0.25 0.00 0.20
SpeciesLeucanthemum_ircutianum 2.394 1.547 0.20 0.00 0.30
SpeciesLigusticum_mutellina 2.394 1.547 0.55 0.00 0.20
SpeciesLinum_catharticum 2.394 1.547 0.31 0.00 0.20
SpeciesLotus_corniculatus 2.394 1.547 0.20 0.00 0.78
SpeciesLuzula_glabrata 2.394 1.547 0.09 0.00 0.09
SpeciesLuzula_multiflora 2.394 1.547 0.28 0.00 0.20
SpeciesLuzula_sylvatica 2.394 1.547 0.28 0.00 0.20
SpeciesLysimachia_nemorum 2.394 1.547 0.09 0.00 0.09
SpeciesMaianthemum_bifolium 2.394 1.547 0.28 0.00 0.20
SpeciesMentha_longifolia 2.394 1.547 0.00 0.17 0.00
SpeciesMinuartia_sedoides 2.394 1.547 0.20 0.00 0.30
SpeciesMoehringia_ciliata 2.394 1.547 0.31 0.00 0.20
SpeciesMyosotis_alpestris 2.394 1.547 0.31 0.00 0.20
SpeciesMyosotis_sylvatica 2.394 1.547 0.55 0.00 0.20
SpeciesNardus_stricta 2.394 1.547 0.00 0.88 0.00
SpeciesOriganum_vulgare 2.394 1.547 0.40 0.00 0.20
SpeciesParnassia_palustris 2.394 1.547 0.28 0.00 0.20
SpeciesPedicularis_rostratocapitata 2.394 1.547 0.09 0.00 0.09
SpeciesPhleum_pratense 2.394 1.547 0.20 0.00 0.21
SpeciesPhyteuma_orbiculare 2.394 1.547 0.25 0.00 0.20
SpeciesPimpinella_major 2.394 1.547 0.28 0.00 0.20
SpeciesPinguicula_alpina 2.394 1.547 0.55 0.00 0.20
SpeciesPlantago_lanceolata 2.394 1.547 0.20 0.00 0.34
SpeciesPlantago_major 2.394 1.547 0.31 0.00 0.20
SpeciesPlantago_media 2.394 1.547 0.31 0.00 0.20
SpeciesPoa_alpina 2.394 1.547 0.31 0.00 0.20
SpeciesPoa_pratensis 2.394 1.547 0.00 0.88 0.00
SpeciesPoa_trivialis 2.394 1.547 0.00 0.88 0.00
SpeciesPolygala_amara 2.394 1.547 0.31 0.00 0.20
SpeciesPolygala_chamaebuxus 2.394 1.547 0.31 0.00 0.20
SpeciesPolygonatum_verticillatum 2.394 1.547 0.28 0.00 0.20
SpeciesPotentilla_aurea 2.394 1.547 0.87 0.00 0.20
SpeciesPotentilla_erecta 2.394 1.547 0.36 0.00 0.20
SpeciesPrimula_auricula 2.394 1.547 0.00 0.88 0.00
SpeciesPrimula_elatior 2.394 1.547 0.25 0.00 0.20
SpeciesPrimula_farinosa 2.394 1.547 0.25 0.00 0.20
SpeciesPrimula_minima 2.394 1.547 0.85 0.00 0.20
SpeciesPrunella_vulgaris 2.394 1.547 0.31 0.00 0.20
SpeciesRanunculus_acris 2.394 1.547 0.00 0.97 0.00
SpeciesRanunculus_alpestris 2.394 1.547 0.65 0.00 0.20
SpeciesRanunculus_montanus 2.394 1.547 0.28 0.00 0.20
SpeciesRanunculus_nemorosus 2.394 1.547 0.20 0.00 0.79
SpeciesRanunculus_repens 2.394 1.547 0.40 0.00 0.20
SpeciesRhinanthus_glacialis 2.394 1.547 0.31 0.00 0.20
SpeciesRhodothamnus_chamaecistus 2.394 1.547 0.31 0.00 0.20
SpeciesRumex_acetosa 2.394 1.547 0.36 0.00 0.20
SpeciesSalix_retusa 2.394 1.547 0.31 0.00 0.20
SpeciesSaxifraga_caesia 2.394 1.547 0.22 0.00 0.20
SpeciesScabiosa_lucida 2.394 1.547 0.40 0.00 0.20
SpeciesSenecio_abrotanifolius 2.394 1.547 0.25 0.00 0.20
SpeciesSesleria_albicans 2.394 1.547 0.25 0.00 0.20
SpeciesSilene_acaulis 2.394 1.547 0.20 0.00 0.30
SpeciesSoldanella_alpina 2.394 1.547 0.09 0.00 0.09
SpeciesSolidago_virgaurea 2.394 1.547 0.00 0.93 0.00
SpeciesStellaria_graminea 2.394 1.547 0.00 0.45 0.00
SpeciesThesium_alpinum 2.394 1.547 0.00 0.69 0.00
SpeciesThymus_pulegioides 2.394 1.547 0.28 0.00 0.20
SpeciesTofieldia_calyculata 2.394 1.547 0.00 0.88 0.00
SpeciesTofieldia_pusilla 2.394 1.547 0.55 0.00 0.20
SpeciesTrifolium_pratense 2.394 1.547 0.31 0.00 0.20
SpeciesTrifolium_repens 2.394 1.547 0.00 0.88 0.00
SpeciesTrollius_europaeus 2.394 1.547 0.63 0.00 0.20
SpeciesVaccinium_myrtillus 2.394 1.547 0.31 0.00 0.20
SpeciesVaccinium_vitis-idaea 2.394 1.547 0.31 0.00 0.20
SpeciesValeriana_saxatilis 2.394 1.547 0.28 0.00 0.20
SpeciesVeratrum_album 2.394 1.547 0.31 0.00 0.20
SpeciesVeronica_aphylla 2.394 1.547 0.20 0.00 0.34
SpeciesVeronica_chamaedrys 2.394 1.547 0.00 0.11 0.00
SpeciesVicia_sepium 2.394 1.547 0.90 0.00 0.20
SpeciesViola_biflora 2.394 1.547 0.40 0.00 0.20
SpeciesWillemetia_stipitata 2.394 1.547 0.09 0.00 0.09
Residual NA NA
0.28
0.00 0.00
0.20 0.20 0.00
0.28 0.81 0.00 0.20
0.28 0.85 0.00 0.20 0.81
0.20 0.20 0.00 0.26 0.20 0.20
0.25 0.25 0.00 0.20 0.25 0.25 0.20
0.00 0.00 0.93 0.00 0.00 0.00 0.00 0.00
0.28 0.48 0.00 0.20 0.48 0.48 0.20 0.25 0.00
0.28 0.77 0.00 0.20 0.77 0.77 0.20 0.25 0.00 0.48
0.00 0.00 0.45 0.00 0.00 0.00 0.00 0.00 0.45 0.00 0.00
0.00 0.00 0.45 0.00 0.00 0.00 0.00 0.00 0.45 0.00 0.00 0.91
0.00 0.00 0.45 0.00 0.00 0.00 0.00 0.00 0.45 0.00 0.00 0.92 0.91
0.00 0.00 0.45 0.00 0.00 0.00 0.00 0.00 0.45 0.00 0.00 0.92 0.91 0.92
0.00 0.00 0.45 0.00 0.00 0.00 0.00 0.00 0.45 0.00 0.00 0.92 0.91 0.92 0.92
0.00 0.00 0.45 0.00 0.00 0.00 0.00 0.00 0.45 0.00 0.00 0.92 0.91 0.92 0.93
0.00 0.00 0.45 0.00 0.00 0.00 0.00 0.00 0.45 0.00 0.00 0.92 0.91 0.92 0.92
0.00 0.00 0.45 0.00 0.00 0.00 0.00 0.00 0.45 0.00 0.00 0.91 0.94 0.91 0.91
0.00 0.00 0.45 0.00 0.00 0.00 0.00 0.00 0.45 0.00 0.00 0.92 0.91 0.92 0.96
0.28 0.77 0.00 0.20 0.77 0.77 0.20 0.25 0.00 0.48 0.83 0.00 0.00 0.00 0.00
0.28 0.77 0.00 0.20 0.77 0.77 0.20 0.25 0.00 0.48 0.90 0.00 0.00 0.00 0.00
0.28 0.81 0.00 0.20 0.87 0.81 0.20 0.25 0.00 0.48 0.77 0.00 0.00 0.00 0.00
0.00 0.00 0.88 0.00 0.00 0.00 0.00 0.00 0.88 0.00 0.00 0.45 0.45 0.45 0.45
0.00 0.00 0.88 0.00 0.00 0.00 0.00 0.00 0.88 0.00 0.00 0.45 0.45 0.45 0.45
0.20 0.20 0.00 0.34 0.20 0.20 0.26 0.20 0.00 0.20 0.20 0.00 0.00 0.00 0.00
0.20 0.20 0.00 0.30 0.20 0.20 0.26 0.20 0.00 0.20 0.20 0.00 0.00 0.00 0.00
0.28 0.31 0.00 0.20 0.31 0.31 0.20 0.25 0.00 0.31 0.31 0.00 0.00 0.00 0.00
0.00 0.00 0.88 0.00 0.00 0.00 0.00 0.00 0.88 0.00 0.00 0.45 0.45 0.45 0.45
0.28 0.31 0.00 0.20 0.31 0.31 0.20 0.25 0.00 0.31 0.31 0.00 0.00 0.00 0.00
0.28 0.31 0.00 0.20 0.31 0.31 0.20 0.25 0.00 0.31 0.31 0.00 0.00 0.00 0.00
0.20 0.20 0.00 0.26 0.20 0.20 0.38 0.20 0.00 0.20 0.20 0.00 0.00 0.00 0.00
0.20 0.20 0.00 0.84 0.20 0.20 0.26 0.20 0.00 0.20 0.20 0.00 0.00 0.00 0.00
0.28 0.83 0.00 0.20 0.81 0.83 0.20 0.25 0.00 0.48 0.77 0.00 0.00 0.00 0.00
0.20 0.20 0.00 0.30 0.20 0.20 0.26 0.20 0.00 0.20 0.20 0.00 0.00 0.00 0.00
0.28 0.81 0.00 0.20 0.87 0.81 0.20 0.25 0.00 0.48 0.77 0.00 0.00 0.00 0.00
0.28 0.37 0.00 0.20 0.37 0.37 0.20 0.25 0.00 0.37 0.37 0.00 0.00 0.00 0.00
0.20 0.20 0.00 0.30 0.20 0.20 0.26 0.20 0.00 0.20 0.20 0.00 0.00 0.00 0.00
0.20 0.20 0.00 0.84 0.20 0.20 0.26 0.20 0.00 0.20 0.20 0.00 0.00 0.00 0.00
0.00 0.00 0.17 0.00 0.00 0.00 0.00 0.00 0.17 0.00 0.00 0.17 0.17 0.17 0.17
0.28 0.31 0.00 0.20 0.31 0.31 0.20 0.25 0.00 0.31 0.31 0.00 0.00 0.00 0.00
0.28 0.37 0.00 0.20 0.37 0.37 0.20 0.25 0.00 0.37 0.37 0.00 0.00 0.00 0.00
0.28 0.31 0.00 0.20 0.31 0.31 0.20 0.25 0.00 0.31 0.31 0.00 0.00 0.00 0.00
0.28 0.31 0.00 0.20 0.31 0.31 0.20 0.25 0.00 0.31 0.31 0.00 0.00 0.00 0.00
0.28 0.31 0.00 0.20 0.31 0.31 0.20 0.25 0.00 0.31 0.31 0.00 0.00 0.00 0.00
0.28 0.31 0.00 0.20 0.31 0.31 0.20 0.25 0.00 0.31 0.31 0.00 0.00 0.00 0.00
0.00 0.00 0.88 0.00 0.00 0.00 0.00 0.00 0.88 0.00 0.00 0.45 0.45 0.45 0.45
0.00 0.00 0.88 0.00 0.00 0.00 0.00 0.00 0.88 0.00 0.00 0.45 0.45 0.45 0.45
0.00 0.00 0.88 0.00 0.00 0.00 0.00 0.00 0.88 0.00 0.00 0.45 0.45 0.45 0.45
0.20 0.20 0.00 0.43 0.20 0.20 0.26 0.20 0.00 0.20 0.20 0.00 0.00 0.00 0.00
0.20 0.20 0.00 0.43 0.20 0.20 0.26 0.20 0.00 0.20 0.20 0.00 0.00 0.00 0.00
0.00 0.00 0.17 0.00 0.00 0.00 0.00 0.00 0.17 0.00 0.00 0.17 0.17 0.17 0.17
0.20 0.20 0.00 0.34 0.20 0.20 0.26 0.20 0.00 0.20 0.20 0.00 0.00 0.00 0.00
0.72 0.28 0.00 0.20 0.28 0.28 0.20 0.25 0.00 0.28 0.28 0.00 0.00 0.00 0.00
0.72 0.28 0.00 0.20 0.28 0.28 0.20 0.25 0.00 0.28 0.28 0.00 0.00 0.00 0.00
0.28 0.31 0.00 0.20 0.31 0.31 0.20 0.25 0.00 0.31 0.31 0.00 0.00 0.00 0.00
0.09 0.09 0.00 0.09 0.09 0.09 0.09 0.09 0.00 0.09 0.09 0.00 0.00 0.00 0.00
0.09 0.09 0.00 0.09 0.09 0.09 0.09 0.09 0.00 0.09 0.09 0.00 0.00 0.00 0.00
0.09 0.09 0.00 0.09 0.09 0.09 0.09 0.09 0.00 0.09 0.09 0.00 0.00 0.00 0.00
0.28 0.37 0.00 0.20 0.37 0.37 0.20 0.25 0.00 0.37 0.37 0.00 0.00 0.00 0.00
0.00 0.00 0.88 0.00 0.00 0.00 0.00 0.00 0.88 0.00 0.00 0.45 0.45 0.45 0.45
0.00 0.00 0.11 0.00 0.00 0.00 0.00 0.00 0.11 0.00 0.00 0.11 0.11 0.11 0.11
0.20 0.20 0.00 0.66 0.20 0.20 0.26 0.20 0.00 0.20 0.20 0.00 0.00 0.00 0.00
0.20 0.20 0.00 0.66 0.20 0.20 0.26 0.20 0.00 0.20 0.20 0.00 0.00 0.00 0.00
0.28 0.37 0.00 0.20 0.37 0.37 0.20 0.25 0.00 0.37 0.37 0.00 0.00 0.00 0.00
0.28 0.31 0.00 0.20 0.31 0.31 0.20 0.25 0.00 0.31 0.31 0.00 0.00 0.00 0.00
0.28 0.31 0.00 0.20 0.31 0.31 0.20 0.25 0.00 0.31 0.31 0.00 0.00 0.00 0.00
0.28 0.84 0.00 0.20 0.81 0.84 0.20 0.25 0.00 0.48 0.77 0.00 0.00 0.00 0.00
0.00 0.00 0.93 0.00 0.00 0.00 0.00 0.00 0.93 0.00 0.00 0.45 0.45 0.45 0.45
0.00 0.00 0.45 0.00 0.00 0.00 0.00 0.00 0.45 0.00 0.00 0.92 0.91 0.92 0.92
0.28 0.37 0.00 0.20 0.37 0.37 0.20 0.25 0.00 0.37 0.37 0.00 0.00 0.00 0.00
0.00 0.00 0.88 0.00 0.00 0.00 0.00 0.00 0.88 0.00 0.00 0.45 0.45 0.45 0.45
0.28 0.31 0.00 0.20 0.31 0.31 0.20 0.25 0.00 0.31 0.31 0.00 0.00 0.00 0.00
0.20 0.20 0.00 0.26 0.20 0.20 0.27 0.20 0.00 0.20 0.20 0.00 0.00 0.00 0.00
0.00 0.00 0.45 0.00 0.00 0.00 0.00 0.00 0.45 0.00 0.00 0.50 0.50 0.50 0.50
0.00 0.00 0.45 0.00 0.00 0.00 0.00 0.00 0.45 0.00 0.00 0.50 0.50 0.50 0.50
0.00 0.00 0.45 0.00 0.00 0.00 0.00 0.00 0.45 0.00 0.00 0.50 0.50 0.50 0.50
0.25 0.25 0.00 0.20 0.25 0.25 0.20 0.43 0.00 0.25 0.25 0.00 0.00 0.00 0.00
0.20 0.20 0.00 0.30 0.20 0.20 0.26 0.20 0.00 0.20 0.20 0.00 0.00 0.00 0.00
0.28 0.31 0.00 0.20 0.31 0.31 0.20 0.25 0.00 0.31 0.31 0.00 0.00 0.00 0.00
0.28 0.48 0.00 0.20 0.48 0.48 0.20 0.25 0.00 0.90 0.48 0.00 0.00 0.00 0.00
0.20 0.20 0.00 0.34 0.20 0.20 0.26 0.20 0.00 0.20 0.20 0.00 0.00 0.00 0.00
0.09 0.09 0.00 0.09 0.09 0.09 0.09 0.09 0.00 0.09 0.09 0.00 0.00 0.00 0.00
0.34 0.28 0.00 0.20 0.28 0.28 0.20 0.25 0.00 0.28 0.28 0.00 0.00 0.00 0.00
0.72 0.28 0.00 0.20 0.28 0.28 0.20 0.25 0.00 0.28 0.28 0.00 0.00 0.00 0.00
0.09 0.09 0.00 0.09 0.09 0.09 0.09 0.09 0.00 0.09 0.09 0.00 0.00 0.00 0.00
0.34 0.28 0.00 0.20 0.28 0.28 0.20 0.25 0.00 0.28 0.28 0.00 0.00 0.00 0.00
0.00 0.00 0.17 0.00 0.00 0.00 0.00 0.00 0.17 0.00 0.00 0.17 0.17 0.17 0.17
0.20 0.20 0.00 0.30 0.20 0.20 0.26 0.20 0.00 0.20 0.20 0.00 0.00 0.00 0.00
0.28 0.85 0.00 0.20 0.81 0.86 0.20 0.25 0.00 0.48 0.77 0.00 0.00 0.00 0.00
0.28 0.85 0.00 0.20 0.81 0.86 0.20 0.25 0.00 0.48 0.77 0.00 0.00 0.00 0.00
0.28 0.31 0.00 0.20 0.31 0.31 0.20 0.25 0.00 0.31 0.31 0.00 0.00 0.00 0.00
0.00 0.00 0.88 0.00 0.00 0.00 0.00 0.00 0.88 0.00 0.00 0.45 0.45 0.45 0.45
0.28 0.31 0.00 0.20 0.31 0.31 0.20 0.25 0.00 0.31 0.31 0.00 0.00 0.00 0.00
0.72 0.28 0.00 0.20 0.28 0.28 0.20 0.25 0.00 0.28 0.28 0.00 0.00 0.00 0.00
0.09 0.09 0.00 0.09 0.09 0.09 0.09 0.09 0.00 0.09 0.09 0.00 0.00 0.00 0.00
0.20 0.20 0.00 0.21 0.20 0.20 0.21 0.20 0.00 0.20 0.20 0.00 0.00 0.00 0.00
0.25 0.25 0.00 0.20 0.25 0.25 0.20 0.43 0.00 0.25 0.25 0.00 0.00 0.00 0.00
0.34 0.28 0.00 0.20 0.28 0.28 0.20 0.25 0.00 0.28 0.28 0.00 0.00 0.00 0.00
0.28 0.31 0.00 0.20 0.31 0.31 0.20 0.25 0.00 0.31 0.31 0.00 0.00 0.00 0.00
0.20 0.20 0.00 0.66 0.20 0.20 0.26 0.20 0.00 0.20 0.20 0.00 0.00 0.00 0.00
0.28 0.85 0.00 0.20 0.81 0.86 0.20 0.25 0.00 0.48 0.77 0.00 0.00 0.00 0.00
0.28 0.83 0.00 0.20 0.81 0.83 0.20 0.25 0.00 0.48 0.77 0.00 0.00 0.00 0.00
0.28 0.85 0.00 0.20 0.81 0.95 0.20 0.25 0.00 0.48 0.77 0.00 0.00 0.00 0.00
0.00 0.00 0.88 0.00 0.00 0.00 0.00 0.00 0.88 0.00 0.00 0.45 0.45 0.45 0.45
0.00 0.00 0.88 0.00 0.00 0.00 0.00 0.00 0.88 0.00 0.00 0.45 0.45 0.45 0.45
0.28 0.37 0.00 0.20 0.37 0.37 0.20 0.25 0.00 0.37 0.37 0.00 0.00 0.00 0.00
0.28 0.85 0.00 0.20 0.81 0.86 0.20 0.25 0.00 0.48 0.77 0.00 0.00 0.00 0.00
0.68 0.28 0.00 0.20 0.28 0.28 0.20 0.25 0.00 0.28 0.28 0.00 0.00 0.00 0.00
0.28 0.31 0.00 0.20 0.31 0.31 0.20 0.25 0.00 0.31 0.31 0.00 0.00 0.00 0.00
0.28 0.31 0.00 0.20 0.31 0.31 0.20 0.25 0.00 0.31 0.31 0.00 0.00 0.00 0.00
0.00 0.00 0.88 0.00 0.00 0.00 0.00 0.00 0.88 0.00 0.00 0.45 0.45 0.45 0.45
0.25 0.25 0.00 0.20 0.25 0.25 0.20 0.77 0.00 0.25 0.25 0.00 0.00 0.00 0.00
0.25 0.25 0.00 0.20 0.25 0.25 0.20 0.43 0.00 0.25 0.25 0.00 0.00 0.00 0.00
0.28 0.31 0.00 0.20 0.31 0.31 0.20 0.25 0.00 0.31 0.31 0.00 0.00 0.00 0.00
0.28 0.81 0.00 0.20 0.87 0.81 0.20 0.25 0.00 0.48 0.77 0.00 0.00 0.00 0.00
0.00 0.00 0.93 0.00 0.00 0.00 0.00 0.00 0.93 0.00 0.00 0.45 0.45 0.45 0.45
0.28 0.31 0.00 0.20 0.31 0.31 0.20 0.25 0.00 0.31 0.31 0.00 0.00 0.00 0.00
0.34 0.28 0.00 0.20 0.28 0.28 0.20 0.25 0.00 0.28 0.28 0.00 0.00 0.00 0.00
0.20 0.20 0.00 0.34 0.20 0.20 0.26 0.20 0.00 0.20 0.20 0.00 0.00 0.00 0.00
0.28 0.31 0.00 0.20 0.31 0.31 0.20 0.25 0.00 0.31 0.31 0.00 0.00 0.00 0.00
0.28 0.81 0.00 0.20 0.86 0.81 0.20 0.25 0.00 0.48 0.77 0.00 0.00 0.00 0.00
0.28 0.37 0.00 0.20 0.37 0.37 0.20 0.25 0.00 0.37 0.37 0.00 0.00 0.00 0.00
0.28 0.31 0.00 0.20 0.31 0.31 0.20 0.25 0.00 0.31 0.31 0.00 0.00 0.00 0.00
0.28 0.85 0.00 0.20 0.81 0.91 0.20 0.25 0.00 0.48 0.77 0.00 0.00 0.00 0.00
0.22 0.22 0.00 0.20 0.22 0.22 0.20 0.22 0.00 0.22 0.22 0.00 0.00 0.00 0.00
0.28 0.31 0.00 0.20 0.31 0.31 0.20 0.25 0.00 0.31 0.31 0.00 0.00 0.00 0.00
0.25 0.25 0.00 0.20 0.25 0.25 0.20 0.43 0.00 0.25 0.25 0.00 0.00 0.00 0.00
0.25 0.25 0.00 0.20 0.25 0.25 0.20 0.43 0.00 0.25 0.25 0.00 0.00 0.00 0.00
0.20 0.20 0.00 0.30 0.20 0.20 0.26 0.20 0.00 0.20 0.20 0.00 0.00 0.00 0.00
0.09 0.09 0.00 0.09 0.09 0.09 0.09 0.09 0.00 0.09 0.09 0.00 0.00 0.00 0.00
0.00 0.00 0.98 0.00 0.00 0.00 0.00 0.00 0.93 0.00 0.00 0.45 0.45 0.45 0.45
0.00 0.00 0.45 0.00 0.00 0.00 0.00 0.00 0.45 0.00 0.00 0.50 0.50 0.50 0.50
0.00 0.00 0.69 0.00 0.00 0.00 0.00 0.00 0.69 0.00 0.00 0.45 0.45 0.45 0.45
0.72 0.28 0.00 0.20 0.28 0.28 0.20 0.25 0.00 0.28 0.28 0.00 0.00 0.00 0.00
0.00 0.00 0.88 0.00 0.00 0.00 0.00 0.00 0.88 0.00 0.00 0.45 0.45 0.45 0.45
0.28 0.31 0.00 0.20 0.31 0.31 0.20 0.25 0.00 0.31 0.31 0.00 0.00 0.00 0.00
0.28 0.81 0.00 0.20 0.87 0.81 0.20 0.25 0.00 0.48 0.77 0.00 0.00 0.00 0.00
0.00 0.00 0.88 0.00 0.00 0.00 0.00 0.00 0.88 0.00 0.00 0.45 0.45 0.45 0.45
0.28 0.31 0.00 0.20 0.31 0.31 0.20 0.25 0.00 0.31 0.31 0.00 0.00 0.00 0.00
0.28 0.83 0.00 0.20 0.81 0.83 0.20 0.25 0.00 0.48 0.77 0.00 0.00 0.00 0.00
0.28 0.81 0.00 0.20 0.87 0.81 0.20 0.25 0.00 0.48 0.77 0.00 0.00 0.00 0.00
0.34 0.28 0.00 0.20 0.28 0.28 0.20 0.25 0.00 0.28 0.28 0.00 0.00 0.00 0.00
0.28 0.81 0.00 0.20 0.87 0.81 0.20 0.25 0.00 0.48 0.77 0.00 0.00 0.00 0.00
0.20 0.20 0.00 0.66 0.20 0.20 0.26 0.20 0.00 0.20 0.20 0.00 0.00 0.00 0.00
0.00 0.00 0.11 0.00 0.00 0.00 0.00 0.00 0.11 0.00 0.00 0.11 0.11 0.11 0.11
0.28 0.31 0.00 0.20 0.31 0.31 0.20 0.25 0.00 0.31 0.31 0.00 0.00 0.00 0.00
0.28 0.31 0.00 0.20 0.31 0.31 0.20 0.25 0.00 0.31 0.31 0.00 0.00 0.00 0.00
0.09 0.09 0.00 0.09 0.09 0.09 0.09 0.09 0.00 0.09 0.09 0.00 0.00 0.00 0.00
0.92
0.92 0.92
0.91 0.91 0.91
0.92 0.93 0.92 0.91
0.00 0.00 0.00 0.00 0.00
0.00 0.00 0.00 0.00 0.00 0.83
0.00 0.00 0.00 0.00 0.00 0.77 0.77
0.45 0.45 0.45 0.45 0.45 0.00 0.00 0.00
0.45 0.45 0.45 0.45 0.45 0.00 0.00 0.00 0.93
0.00 0.00 0.00 0.00 0.00 0.20 0.20 0.20 0.00 0.00
0.00 0.00 0.00 0.00 0.00 0.20 0.20 0.20 0.00 0.00 0.30
0.00 0.00 0.00 0.00 0.00 0.31 0.31 0.31 0.00 0.00 0.20 0.20
0.45 0.45 0.45 0.45 0.45 0.00 0.00 0.00 0.91 0.91 0.00 0.00 0.00
0.00 0.00 0.00 0.00 0.00 0.31 0.31 0.31 0.00 0.00 0.20 0.20 0.40 0.00
0.00 0.00 0.00 0.00 0.00 0.31 0.31 0.31 0.00 0.00 0.20 0.20 0.40 0.00 0.53
0.00 0.00 0.00 0.00 0.00 0.20 0.20 0.20 0.00 0.00 0.26 0.26 0.20 0.00 0.20
0.00 0.00 0.00 0.00 0.00 0.20 0.20 0.20 0.00 0.00 0.34 0.30 0.20 0.00 0.20
0.00 0.00 0.00 0.00 0.00 0.77 0.77 0.81 0.00 0.00 0.20 0.20 0.31 0.00 0.31
0.00 0.00 0.00 0.00 0.00 0.20 0.20 0.20 0.00 0.00 0.30 0.34 0.20 0.00 0.20
0.00 0.00 0.00 0.00 0.00 0.77 0.77 0.88 0.00 0.00 0.20 0.20 0.31 0.00 0.31
0.00 0.00 0.00 0.00 0.00 0.37 0.37 0.37 0.00 0.00 0.20 0.20 0.31 0.00 0.31
0.00 0.00 0.00 0.00 0.00 0.20 0.20 0.20 0.00 0.00 0.30 0.34 0.20 0.00 0.20
0.00 0.00 0.00 0.00 0.00 0.20 0.20 0.20 0.00 0.00 0.34 0.30 0.20 0.00 0.20
0.17 0.17 0.17 0.17 0.17 0.00 0.00 0.00 0.17 0.17 0.00 0.00 0.00 0.17 0.00
0.00 0.00 0.00 0.00 0.00 0.31 0.31 0.31 0.00 0.00 0.20 0.20 0.55 0.00 0.40
0.00 0.00 0.00 0.00 0.00 0.37 0.37 0.37 0.00 0.00 0.20 0.20 0.31 0.00 0.31
0.00 0.00 0.00 0.00 0.00 0.31 0.31 0.31 0.00 0.00 0.20 0.20 0.55 0.00 0.40
0.00 0.00 0.00 0.00 0.00 0.31 0.31 0.31 0.00 0.00 0.20 0.20 0.55 0.00 0.40
0.00 0.00 0.00 0.00 0.00 0.31 0.31 0.31 0.00 0.00 0.20 0.20 0.55 0.00 0.40
0.00 0.00 0.00 0.00 0.00 0.31 0.31 0.31 0.00 0.00 0.20 0.20 0.55 0.00 0.40
0.45 0.45 0.45 0.45 0.45 0.00 0.00 0.00 0.90 0.90 0.00 0.00 0.00 0.90 0.00
0.45 0.45 0.45 0.45 0.45 0.00 0.00 0.00 0.90 0.90 0.00 0.00 0.00 0.90 0.00
0.45 0.45 0.45 0.45 0.45 0.00 0.00 0.00 0.90 0.90 0.00 0.00 0.00 0.90 0.00
0.00 0.00 0.00 0.00 0.00 0.20 0.20 0.20 0.00 0.00 0.34 0.30 0.20 0.00 0.20
0.00 0.00 0.00 0.00 0.00 0.20 0.20 0.20 0.00 0.00 0.34 0.30 0.20 0.00 0.20
0.17 0.17 0.17 0.17 0.17 0.00 0.00 0.00 0.17 0.17 0.00 0.00 0.00 0.17 0.00
0.00 0.00 0.00 0.00 0.00 0.20 0.20 0.20 0.00 0.00 0.51 0.30 0.20 0.00 0.20
0.00 0.00 0.00 0.00 0.00 0.28 0.28 0.28 0.00 0.00 0.20 0.20 0.28 0.00 0.28
0.00 0.00 0.00 0.00 0.00 0.28 0.28 0.28 0.00 0.00 0.20 0.20 0.28 0.00 0.28
0.00 0.00 0.00 0.00 0.00 0.31 0.31 0.31 0.00 0.00 0.20 0.20 0.55 0.00 0.40
0.00 0.00 0.00 0.00 0.00 0.09 0.09 0.09 0.00 0.00 0.09 0.09 0.09 0.00 0.09
0.00 0.00 0.00 0.00 0.00 0.09 0.09 0.09 0.00 0.00 0.09 0.09 0.09 0.00 0.09
0.00 0.00 0.00 0.00 0.00 0.09 0.09 0.09 0.00 0.00 0.09 0.09 0.09 0.00 0.09
0.00 0.00 0.00 0.00 0.00 0.37 0.37 0.37 0.00 0.00 0.20 0.20 0.31 0.00 0.31
0.45 0.45 0.45 0.45 0.45 0.00 0.00 0.00 0.91 0.91 0.00 0.00 0.00 0.91 0.00
0.11 0.11 0.11 0.11 0.11 0.00 0.00 0.00 0.11 0.11 0.00 0.00 0.00 0.11 0.00
0.00 0.00 0.00 0.00 0.00 0.20 0.20 0.20 0.00 0.00 0.34 0.30 0.20 0.00 0.20
0.00 0.00 0.00 0.00 0.00 0.20 0.20 0.20 0.00 0.00 0.34 0.30 0.20 0.00 0.20
0.00 0.00 0.00 0.00 0.00 0.37 0.37 0.37 0.00 0.00 0.20 0.20 0.31 0.00 0.31
0.00 0.00 0.00 0.00 0.00 0.31 0.31 0.31 0.00 0.00 0.20 0.20 0.55 0.00 0.40
0.00 0.00 0.00 0.00 0.00 0.31 0.31 0.31 0.00 0.00 0.20 0.20 0.85 0.00 0.40
0.00 0.00 0.00 0.00 0.00 0.77 0.77 0.81 0.00 0.00 0.20 0.20 0.31 0.00 0.31
0.45 0.45 0.45 0.45 0.45 0.00 0.00 0.00 0.88 0.88 0.00 0.00 0.00 0.88 0.00
0.99 0.92 0.92 0.91 0.92 0.00 0.00 0.00 0.45 0.45 0.00 0.00 0.00 0.45 0.00
0.00 0.00 0.00 0.00 0.00 0.37 0.37 0.37 0.00 0.00 0.20 0.20 0.31 0.00 0.31
0.45 0.45 0.45 0.45 0.45 0.00 0.00 0.00 0.91 0.91 0.00 0.00 0.00 0.91 0.00
0.00 0.00 0.00 0.00 0.00 0.31 0.31 0.31 0.00 0.00 0.20 0.20 0.40 0.00 0.53
0.00 0.00 0.00 0.00 0.00 0.20 0.20 0.20 0.00 0.00 0.26 0.26 0.20 0.00 0.20
0.50 0.50 0.50 0.50 0.50 0.00 0.00 0.00 0.45 0.45 0.00 0.00 0.00 0.45 0.00
0.50 0.50 0.50 0.50 0.50 0.00 0.00 0.00 0.45 0.45 0.00 0.00 0.00 0.45 0.00
0.50 0.50 0.50 0.50 0.50 0.00 0.00 0.00 0.45 0.45 0.00 0.00 0.00 0.45 0.00
0.00 0.00 0.00 0.00 0.00 0.25 0.25 0.25 0.00 0.00 0.20 0.20 0.25 0.00 0.25
0.00 0.00 0.00 0.00 0.00 0.20 0.20 0.20 0.00 0.00 0.30 0.31 0.20 0.00 0.20
0.00 0.00 0.00 0.00 0.00 0.31 0.31 0.31 0.00 0.00 0.20 0.20 0.77 0.00 0.40
0.00 0.00 0.00 0.00 0.00 0.48 0.48 0.48 0.00 0.00 0.20 0.20 0.31 0.00 0.31
0.00 0.00 0.00 0.00 0.00 0.20 0.20 0.20 0.00 0.00 0.51 0.30 0.20 0.00 0.20
0.00 0.00 0.00 0.00 0.00 0.09 0.09 0.09 0.00 0.00 0.09 0.09 0.09 0.00 0.09
0.00 0.00 0.00 0.00 0.00 0.28 0.28 0.28 0.00 0.00 0.20 0.20 0.28 0.00 0.28
0.00 0.00 0.00 0.00 0.00 0.28 0.28 0.28 0.00 0.00 0.20 0.20 0.28 0.00 0.28
0.00 0.00 0.00 0.00 0.00 0.09 0.09 0.09 0.00 0.00 0.09 0.09 0.09 0.00 0.09
0.00 0.00 0.00 0.00 0.00 0.28 0.28 0.28 0.00 0.00 0.20 0.20 0.28 0.00 0.28
0.17 0.17 0.17 0.17 0.17 0.00 0.00 0.00 0.17 0.17 0.00 0.00 0.00 0.17 0.00
0.00 0.00 0.00 0.00 0.00 0.20 0.20 0.20 0.00 0.00 0.30 0.34 0.20 0.00 0.20
0.00 0.00 0.00 0.00 0.00 0.77 0.77 0.81 0.00 0.00 0.20 0.20 0.31 0.00 0.31
0.00 0.00 0.00 0.00 0.00 0.77 0.77 0.81 0.00 0.00 0.20 0.20 0.31 0.00 0.31
0.00 0.00 0.00 0.00 0.00 0.31 0.31 0.31 0.00 0.00 0.20 0.20 0.98 0.00 0.40
0.45 0.45 0.45 0.45 0.45 0.00 0.00 0.00 0.91 0.91 0.00 0.00 0.00 0.91 0.00
0.00 0.00 0.00 0.00 0.00 0.31 0.31 0.31 0.00 0.00 0.20 0.20 0.40 0.00 0.53
0.00 0.00 0.00 0.00 0.00 0.28 0.28 0.28 0.00 0.00 0.20 0.20 0.28 0.00 0.28
0.00 0.00 0.00 0.00 0.00 0.09 0.09 0.09 0.00 0.00 0.09 0.09 0.09 0.00 0.09
0.00 0.00 0.00 0.00 0.00 0.20 0.20 0.20 0.00 0.00 0.21 0.21 0.20 0.00 0.20
0.00 0.00 0.00 0.00 0.00 0.25 0.25 0.25 0.00 0.00 0.20 0.20 0.25 0.00 0.25
0.00 0.00 0.00 0.00 0.00 0.28 0.28 0.28 0.00 0.00 0.20 0.20 0.28 0.00 0.28
0.00 0.00 0.00 0.00 0.00 0.31 0.31 0.31 0.00 0.00 0.20 0.20 0.55 0.00 0.40
0.00 0.00 0.00 0.00 0.00 0.20 0.20 0.20 0.00 0.00 0.34 0.30 0.20 0.00 0.20
0.00 0.00 0.00 0.00 0.00 0.77 0.77 0.81 0.00 0.00 0.20 0.20 0.31 0.00 0.31
0.00 0.00 0.00 0.00 0.00 0.77 0.77 0.81 0.00 0.00 0.20 0.20 0.31 0.00 0.31
0.00 0.00 0.00 0.00 0.00 0.77 0.77 0.81 0.00 0.00 0.20 0.20 0.31 0.00 0.31
0.45 0.45 0.45 0.45 0.45 0.00 0.00 0.00 0.91 0.91 0.00 0.00 0.00 0.91 0.00
0.45 0.45 0.45 0.45 0.45 0.00 0.00 0.00 0.88 0.88 0.00 0.00 0.00 0.88 0.00
0.00 0.00 0.00 0.00 0.00 0.37 0.37 0.37 0.00 0.00 0.20 0.20 0.31 0.00 0.31
0.00 0.00 0.00 0.00 0.00 0.77 0.77 0.81 0.00 0.00 0.20 0.20 0.31 0.00 0.31
0.00 0.00 0.00 0.00 0.00 0.28 0.28 0.28 0.00 0.00 0.20 0.20 0.28 0.00 0.28
0.00 0.00 0.00 0.00 0.00 0.31 0.31 0.31 0.00 0.00 0.20 0.20 0.55 0.00 0.40
0.00 0.00 0.00 0.00 0.00 0.31 0.31 0.31 0.00 0.00 0.20 0.20 0.36 0.00 0.36
0.45 0.45 0.45 0.45 0.45 0.00 0.00 0.00 0.90 0.90 0.00 0.00 0.00 0.90 0.00
0.00 0.00 0.00 0.00 0.00 0.25 0.25 0.25 0.00 0.00 0.20 0.20 0.25 0.00 0.25
0.00 0.00 0.00 0.00 0.00 0.25 0.25 0.25 0.00 0.00 0.20 0.20 0.25 0.00 0.25
0.00 0.00 0.00 0.00 0.00 0.31 0.31 0.31 0.00 0.00 0.20 0.20 0.55 0.00 0.40
0.00 0.00 0.00 0.00 0.00 0.77 0.77 0.88 0.00 0.00 0.20 0.20 0.31 0.00 0.31
0.45 0.45 0.45 0.45 0.45 0.00 0.00 0.00 0.88 0.88 0.00 0.00 0.00 0.88 0.00
0.00 0.00 0.00 0.00 0.00 0.31 0.31 0.31 0.00 0.00 0.20 0.20 0.55 0.00 0.40
0.00 0.00 0.00 0.00 0.00 0.28 0.28 0.28 0.00 0.00 0.20 0.20 0.28 0.00 0.28
0.00 0.00 0.00 0.00 0.00 0.20 0.20 0.20 0.00 0.00 0.51 0.30 0.20 0.00 0.20
0.00 0.00 0.00 0.00 0.00 0.31 0.31 0.31 0.00 0.00 0.20 0.20 0.40 0.00 0.53
0.00 0.00 0.00 0.00 0.00 0.77 0.77 0.86 0.00 0.00 0.20 0.20 0.31 0.00 0.31
0.00 0.00 0.00 0.00 0.00 0.37 0.37 0.37 0.00 0.00 0.20 0.20 0.31 0.00 0.31
0.00 0.00 0.00 0.00 0.00 0.31 0.31 0.31 0.00 0.00 0.20 0.20 0.36 0.00 0.36
0.00 0.00 0.00 0.00 0.00 0.77 0.77 0.81 0.00 0.00 0.20 0.20 0.31 0.00 0.31
0.00 0.00 0.00 0.00 0.00 0.22 0.22 0.22 0.00 0.00 0.20 0.20 0.22 0.00 0.22
0.00 0.00 0.00 0.00 0.00 0.31 0.31 0.31 0.00 0.00 0.20 0.20 0.40 0.00 0.53
0.00 0.00 0.00 0.00 0.00 0.25 0.25 0.25 0.00 0.00 0.20 0.20 0.25 0.00 0.25
0.00 0.00 0.00 0.00 0.00 0.25 0.25 0.25 0.00 0.00 0.20 0.20 0.25 0.00 0.25
0.00 0.00 0.00 0.00 0.00 0.20 0.20 0.20 0.00 0.00 0.30 0.34 0.20 0.00 0.20
0.00 0.00 0.00 0.00 0.00 0.09 0.09 0.09 0.00 0.00 0.09 0.09 0.09 0.00 0.09
0.45 0.45 0.45 0.45 0.45 0.00 0.00 0.00 0.88 0.88 0.00 0.00 0.00 0.88 0.00
0.50 0.50 0.50 0.50 0.50 0.00 0.00 0.00 0.45 0.45 0.00 0.00 0.00 0.45 0.00
0.45 0.45 0.45 0.45 0.45 0.00 0.00 0.00 0.69 0.69 0.00 0.00 0.00 0.69 0.00
0.00 0.00 0.00 0.00 0.00 0.28 0.28 0.28 0.00 0.00 0.20 0.20 0.28 0.00 0.28
0.45 0.45 0.45 0.45 0.45 0.00 0.00 0.00 0.91 0.91 0.00 0.00 0.00 0.91 0.00
0.00 0.00 0.00 0.00 0.00 0.31 0.31 0.31 0.00 0.00 0.20 0.20 0.83 0.00 0.40
0.00 0.00 0.00 0.00 0.00 0.77 0.77 0.97 0.00 0.00 0.20 0.20 0.31 0.00 0.31
0.45 0.45 0.45 0.45 0.45 0.00 0.00 0.00 0.91 0.91 0.00 0.00 0.00 0.91 0.00
0.00 0.00 0.00 0.00 0.00 0.31 0.31 0.31 0.00 0.00 0.20 0.20 0.55 0.00 0.40
0.00 0.00 0.00 0.00 0.00 0.77 0.77 0.81 0.00 0.00 0.20 0.20 0.31 0.00 0.31
0.00 0.00 0.00 0.00 0.00 0.77 0.77 0.98 0.00 0.00 0.20 0.20 0.31 0.00 0.31
0.00 0.00 0.00 0.00 0.00 0.28 0.28 0.28 0.00 0.00 0.20 0.20 0.28 0.00 0.28
0.00 0.00 0.00 0.00 0.00 0.77 0.77 0.94 0.00 0.00 0.20 0.20 0.31 0.00 0.31
0.00 0.00 0.00 0.00 0.00 0.20 0.20 0.20 0.00 0.00 0.34 0.30 0.20 0.00 0.20
0.11 0.11 0.11 0.11 0.11 0.00 0.00 0.00 0.11 0.11 0.00 0.00 0.00 0.11 0.00
0.00 0.00 0.00 0.00 0.00 0.31 0.31 0.31 0.00 0.00 0.20 0.20 0.55 0.00 0.40
0.00 0.00 0.00 0.00 0.00 0.31 0.31 0.31 0.00 0.00 0.20 0.20 0.40 0.00 0.92
0.00 0.00 0.00 0.00 0.00 0.09 0.09 0.09 0.00 0.00 0.09 0.09 0.09 0.00 0.09
0.20
0.20 0.26
0.31 0.20 0.20
0.20 0.26 0.30 0.20
0.31 0.20 0.20 0.81 0.20
0.31 0.20 0.20 0.37 0.20 0.37
0.20 0.26 0.30 0.20 0.34 0.20 0.20
0.20 0.26 0.85 0.20 0.30 0.20 0.20 0.30
0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00
0.40 0.20 0.20 0.31 0.20 0.31 0.31 0.20 0.20 0.00
0.31 0.20 0.20 0.37 0.20 0.37 0.70 0.20 0.20 0.00 0.31
0.40 0.20 0.20 0.31 0.20 0.31 0.31 0.20 0.20 0.00 0.55 0.31
0.40 0.20 0.20 0.31 0.20 0.31 0.31 0.20 0.20 0.00 0.55 0.31 0.55
0.40 0.20 0.20 0.31 0.20 0.31 0.31 0.20 0.20 0.00 0.55 0.31 0.55 0.88
0.40 0.20 0.20 0.31 0.20 0.31 0.31 0.20 0.20 0.00 0.55 0.31 0.55 0.88 0.93
0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.17 0.00 0.00 0.00 0.00 0.00
0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.17 0.00 0.00 0.00 0.00 0.00
0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.17 0.00 0.00 0.00 0.00 0.00
0.20 0.26 0.43 0.20 0.30 0.20 0.20 0.30 0.43 0.00 0.20 0.20 0.20 0.20 0.20
0.20 0.26 0.43 0.20 0.30 0.20 0.20 0.30 0.43 0.00 0.20 0.20 0.20 0.20 0.20
0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.94 0.00 0.00 0.00 0.00 0.00
0.20 0.26 0.34 0.20 0.30 0.20 0.20 0.30 0.34 0.00 0.20 0.20 0.20 0.20 0.20
0.28 0.20 0.20 0.28 0.20 0.28 0.28 0.20 0.20 0.00 0.28 0.28 0.28 0.28 0.28
0.28 0.20 0.20 0.28 0.20 0.28 0.28 0.20 0.20 0.00 0.28 0.28 0.28 0.28 0.28
0.40 0.20 0.20 0.31 0.20 0.31 0.31 0.20 0.20 0.00 0.69 0.31 0.55 0.55 0.55
0.09 0.09 0.09 0.09 0.09 0.09 0.09 0.09 0.09 0.00 0.09 0.09 0.09 0.09 0.09
0.09 0.09 0.09 0.09 0.09 0.09 0.09 0.09 0.09 0.00 0.09 0.09 0.09 0.09 0.09
0.09 0.09 0.09 0.09 0.09 0.09 0.09 0.09 0.09 0.00 0.09 0.09 0.09 0.09 0.09
0.31 0.20 0.20 0.37 0.20 0.37 0.38 0.20 0.20 0.00 0.31 0.38 0.31 0.31 0.31
0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.17 0.00 0.00 0.00 0.00 0.00
0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.11 0.00 0.00 0.00 0.00 0.00
0.20 0.26 0.66 0.20 0.30 0.20 0.20 0.30 0.66 0.00 0.20 0.20 0.20 0.20 0.20
0.20 0.26 0.66 0.20 0.30 0.20 0.20 0.30 0.66 0.00 0.20 0.20 0.20 0.20 0.20
0.31 0.20 0.20 0.37 0.20 0.37 0.38 0.20 0.20 0.00 0.31 0.38 0.31 0.31 0.31
0.40 0.20 0.20 0.31 0.20 0.31 0.31 0.20 0.20 0.00 0.55 0.31 0.55 0.76 0.76
0.40 0.20 0.20 0.31 0.20 0.31 0.31 0.20 0.20 0.00 0.55 0.31 0.55 0.55 0.55
0.31 0.20 0.20 0.83 0.20 0.81 0.37 0.20 0.20 0.00 0.31 0.37 0.31 0.31 0.31
0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.17 0.00 0.00 0.00 0.00 0.00
0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.17 0.00 0.00 0.00 0.00 0.00
0.31 0.20 0.20 0.37 0.20 0.37 0.70 0.20 0.20 0.00 0.31 0.70 0.31 0.31 0.31
0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.17 0.00 0.00 0.00 0.00 0.00
0.86 0.20 0.20 0.31 0.20 0.31 0.31 0.20 0.20 0.00 0.40 0.31 0.40 0.40 0.40
0.20 0.27 0.26 0.20 0.26 0.20 0.20 0.26 0.26 0.00 0.20 0.20 0.20 0.20 0.20
0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.17 0.00 0.00 0.00 0.00 0.00
0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.17 0.00 0.00 0.00 0.00 0.00
0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.17 0.00 0.00 0.00 0.00 0.00
0.25 0.20 0.20 0.25 0.20 0.25 0.25 0.20 0.20 0.00 0.25 0.25 0.25 0.25 0.25
0.20 0.26 0.30 0.20 0.31 0.20 0.20 0.31 0.30 0.00 0.20 0.20 0.20 0.20 0.20
0.40 0.20 0.20 0.31 0.20 0.31 0.31 0.20 0.20 0.00 0.55 0.31 0.55 0.55 0.55
0.31 0.20 0.20 0.48 0.20 0.48 0.37 0.20 0.20 0.00 0.31 0.37 0.31 0.31 0.31
0.20 0.26 0.34 0.20 0.30 0.20 0.20 0.30 0.34 0.00 0.20 0.20 0.20 0.20 0.20
0.09 0.09 0.09 0.09 0.09 0.09 0.09 0.09 0.09 0.00 0.09 0.09 0.09 0.09 0.09
0.28 0.20 0.20 0.28 0.20 0.28 0.28 0.20 0.20 0.00 0.28 0.28 0.28 0.28 0.28
0.28 0.20 0.20 0.28 0.20 0.28 0.28 0.20 0.20 0.00 0.28 0.28 0.28 0.28 0.28
0.09 0.09 0.09 0.09 0.09 0.09 0.09 0.09 0.09 0.00 0.09 0.09 0.09 0.09 0.09
0.28 0.20 0.20 0.28 0.20 0.28 0.28 0.20 0.20 0.00 0.28 0.28 0.28 0.28 0.28
0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.17 0.00 0.00 0.00 0.00 0.00
0.20 0.26 0.30 0.20 0.34 0.20 0.20 0.34 0.30 0.00 0.20 0.20 0.20 0.20 0.20
0.31 0.20 0.20 0.83 0.20 0.81 0.37 0.20 0.20 0.00 0.31 0.37 0.31 0.31 0.31
0.31 0.20 0.20 0.83 0.20 0.81 0.37 0.20 0.20 0.00 0.31 0.37 0.31 0.31 0.31
0.40 0.20 0.20 0.31 0.20 0.31 0.31 0.20 0.20 0.00 0.55 0.31 0.55 0.55 0.55
0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.17 0.00 0.00 0.00 0.00 0.00
0.78 0.20 0.20 0.31 0.20 0.31 0.31 0.20 0.20 0.00 0.40 0.31 0.40 0.40 0.40
0.28 0.20 0.20 0.28 0.20 0.28 0.28 0.20 0.20 0.00 0.28 0.28 0.28 0.28 0.28
0.09 0.09 0.09 0.09 0.09 0.09 0.09 0.09 0.09 0.00 0.09 0.09 0.09 0.09 0.09
0.20 0.21 0.21 0.20 0.21 0.20 0.20 0.21 0.21 0.00 0.20 0.20 0.20 0.20 0.20
0.25 0.20 0.20 0.25 0.20 0.25 0.25 0.20 0.20 0.00 0.25 0.25 0.25 0.25 0.25
0.28 0.20 0.20 0.28 0.20 0.28 0.28 0.20 0.20 0.00 0.28 0.28 0.28 0.28 0.28
0.40 0.20 0.20 0.31 0.20 0.31 0.31 0.20 0.20 0.00 0.55 0.31 0.55 0.76 0.76
0.20 0.26 0.66 0.20 0.30 0.20 0.20 0.30 0.66 0.00 0.20 0.20 0.20 0.20 0.20
0.31 0.20 0.20 0.83 0.20 0.81 0.37 0.20 0.20 0.00 0.31 0.37 0.31 0.31 0.31
0.31 0.20 0.20 0.91 0.20 0.81 0.37 0.20 0.20 0.00 0.31 0.37 0.31 0.31 0.31
0.31 0.20 0.20 0.83 0.20 0.81 0.37 0.20 0.20 0.00 0.31 0.37 0.31 0.31 0.31
0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.17 0.00 0.00 0.00 0.00 0.00
0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.17 0.00 0.00 0.00 0.00 0.00
0.31 0.20 0.20 0.37 0.20 0.37 0.70 0.20 0.20 0.00 0.31 0.76 0.31 0.31 0.31
0.31 0.20 0.20 0.83 0.20 0.81 0.37 0.20 0.20 0.00 0.31 0.37 0.31 0.31 0.31
0.28 0.20 0.20 0.28 0.20 0.28 0.28 0.20 0.20 0.00 0.28 0.28 0.28 0.28 0.28
0.40 0.20 0.20 0.31 0.20 0.31 0.31 0.20 0.20 0.00 0.85 0.31 0.55 0.55 0.55
0.36 0.20 0.20 0.31 0.20 0.31 0.31 0.20 0.20 0.00 0.36 0.31 0.36 0.36 0.36
0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.17 0.00 0.00 0.00 0.00 0.00
0.25 0.20 0.20 0.25 0.20 0.25 0.25 0.20 0.20 0.00 0.25 0.25 0.25 0.25 0.25
0.25 0.20 0.20 0.25 0.20 0.25 0.25 0.20 0.20 0.00 0.25 0.25 0.25 0.25 0.25
0.40 0.20 0.20 0.31 0.20 0.31 0.31 0.20 0.20 0.00 0.95 0.31 0.55 0.55 0.55
0.31 0.20 0.20 0.81 0.20 0.88 0.37 0.20 0.20 0.00 0.31 0.37 0.31 0.31 0.31
0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.17 0.00 0.00 0.00 0.00 0.00
0.40 0.20 0.20 0.31 0.20 0.31 0.31 0.20 0.20 0.00 0.65 0.31 0.55 0.55 0.55
0.28 0.20 0.20 0.28 0.20 0.28 0.28 0.20 0.20 0.00 0.28 0.28 0.28 0.28 0.28
0.20 0.26 0.34 0.20 0.30 0.20 0.20 0.30 0.34 0.00 0.20 0.20 0.20 0.20 0.20
0.89 0.20 0.20 0.31 0.20 0.31 0.31 0.20 0.20 0.00 0.40 0.31 0.40 0.40 0.40
0.31 0.20 0.20 0.81 0.20 0.86 0.37 0.20 0.20 0.00 0.31 0.37 0.31 0.31 0.31
0.31 0.20 0.20 0.37 0.20 0.37 0.38 0.20 0.20 0.00 0.31 0.38 0.31 0.31 0.31
0.36 0.20 0.20 0.31 0.20 0.31 0.31 0.20 0.20 0.00 0.36 0.31 0.36 0.36 0.36
0.31 0.20 0.20 0.83 0.20 0.81 0.37 0.20 0.20 0.00 0.31 0.37 0.31 0.31 0.31
0.22 0.20 0.20 0.22 0.20 0.22 0.22 0.20 0.20 0.00 0.22 0.22 0.22 0.22 0.22
0.86 0.20 0.20 0.31 0.20 0.31 0.31 0.20 0.20 0.00 0.40 0.31 0.40 0.40 0.40
0.25 0.20 0.20 0.25 0.20 0.25 0.25 0.20 0.20 0.00 0.25 0.25 0.25 0.25 0.25
0.25 0.20 0.20 0.25 0.20 0.25 0.25 0.20 0.20 0.00 0.25 0.25 0.25 0.25 0.25
0.20 0.26 0.30 0.20 0.34 0.20 0.20 0.34 0.30 0.00 0.20 0.20 0.20 0.20 0.20
0.09 0.09 0.09 0.09 0.09 0.09 0.09 0.09 0.09 0.00 0.09 0.09 0.09 0.09 0.09
0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.17 0.00 0.00 0.00 0.00 0.00
0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.17 0.00 0.00 0.00 0.00 0.00
0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.17 0.00 0.00 0.00 0.00 0.00
0.28 0.20 0.20 0.28 0.20 0.28 0.28 0.20 0.20 0.00 0.28 0.28 0.28 0.28 0.28
0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.17 0.00 0.00 0.00 0.00 0.00
0.40 0.20 0.20 0.31 0.20 0.31 0.31 0.20 0.20 0.00 0.55 0.31 0.55 0.55 0.55
0.31 0.20 0.20 0.81 0.20 0.88 0.37 0.20 0.20 0.00 0.31 0.37 0.31 0.31 0.31
0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.17 0.00 0.00 0.00 0.00 0.00
0.40 0.20 0.20 0.31 0.20 0.31 0.31 0.20 0.20 0.00 0.63 0.31 0.55 0.55 0.55
0.31 0.20 0.20 0.91 0.20 0.81 0.37 0.20 0.20 0.00 0.31 0.37 0.31 0.31 0.31
0.31 0.20 0.20 0.81 0.20 0.88 0.37 0.20 0.20 0.00 0.31 0.37 0.31 0.31 0.31
0.28 0.20 0.20 0.28 0.20 0.28 0.28 0.20 0.20 0.00 0.28 0.28 0.28 0.28 0.28
0.31 0.20 0.20 0.81 0.20 0.88 0.37 0.20 0.20 0.00 0.31 0.37 0.31 0.31 0.31
0.20 0.26 0.66 0.20 0.30 0.20 0.20 0.30 0.66 0.00 0.20 0.20 0.20 0.20 0.20
0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.11 0.00 0.00 0.00 0.00 0.00
0.40 0.20 0.20 0.31 0.20 0.31 0.31 0.20 0.20 0.00 0.85 0.31 0.55 0.55 0.55
0.53 0.20 0.20 0.31 0.20 0.31 0.31 0.20 0.20 0.00 0.40 0.31 0.40 0.40 0.40
0.09 0.09 0.09 0.09 0.09 0.09 0.09 0.09 0.09 0.00 0.09 0.09 0.09 0.09 0.09
0.00
0.00 0.93
0.00 0.93 0.95
0.20 0.00 0.00 0.00
0.20 0.00 0.00 0.00 0.72
0.00 0.17 0.17 0.17 0.00 0.00
0.20 0.00 0.00 0.00 0.34 0.34 0.00
0.28 0.00 0.00 0.00 0.20 0.20 0.00 0.20
0.28 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.84
0.55 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.28 0.28
0.09 0.00 0.00 0.00 0.09 0.09 0.00 0.09 0.09 0.09 0.09
0.09 0.00 0.00 0.00 0.09 0.09 0.00 0.09 0.09 0.09 0.09 0.92
0.09 0.00 0.00 0.00 0.09 0.09 0.00 0.09 0.09 0.09 0.09 0.92 0.92
0.31 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.28 0.28 0.31 0.09 0.09 0.09
0.00 0.90 0.90 0.90 0.00 0.00 0.17 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00
0.00 0.11 0.11 0.11 0.00 0.00 0.11 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00
0.20 0.00 0.00 0.00 0.43 0.43 0.00 0.34 0.20 0.20 0.20 0.09 0.09 0.09 0.20
0.20 0.00 0.00 0.00 0.43 0.43 0.00 0.34 0.20 0.20 0.20 0.09 0.09 0.09 0.20
0.31 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.28 0.28 0.31 0.09 0.09 0.09 0.59
0.76 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.28 0.28 0.55 0.09 0.09 0.09 0.31
0.55 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.28 0.28 0.55 0.09 0.09 0.09 0.31
0.31 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.28 0.28 0.31 0.09 0.09 0.09 0.37
0.00 0.88 0.88 0.88 0.00 0.00 0.17 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00
0.00 0.45 0.45 0.45 0.00 0.00 0.17 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00
0.31 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.28 0.28 0.31 0.09 0.09 0.09 0.38
0.00 0.90 0.90 0.90 0.00 0.00 0.17 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00
0.40 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.28 0.28 0.40 0.09 0.09 0.09 0.31
0.20 0.00 0.00 0.00 0.26 0.26 0.00 0.26 0.20 0.20 0.20 0.09 0.09 0.09 0.20
0.00 0.45 0.45 0.45 0.00 0.00 0.17 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00
0.00 0.45 0.45 0.45 0.00 0.00 0.17 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00
0.00 0.45 0.45 0.45 0.00 0.00 0.17 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00
0.25 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.25 0.25 0.25 0.09 0.09 0.09 0.25
0.20 0.00 0.00 0.00 0.30 0.30 0.00 0.30 0.20 0.20 0.20 0.09 0.09 0.09 0.20
0.55 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.28 0.28 0.55 0.09 0.09 0.09 0.31
0.31 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.28 0.28 0.31 0.09 0.09 0.09 0.37
0.20 0.00 0.00 0.00 0.34 0.34 0.00 0.88 0.20 0.20 0.20 0.09 0.09 0.09 0.20
0.09 0.00 0.00 0.00 0.09 0.09 0.00 0.09 0.09 0.09 0.09 0.92 0.92 0.99 0.09
0.28 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.34 0.34 0.28 0.09 0.09 0.09 0.28
0.28 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.75 0.75 0.28 0.09 0.09 0.09 0.28
0.09 0.00 0.00 0.00 0.09 0.09 0.00 0.09 0.09 0.09 0.09 0.54 0.54 0.54 0.09
0.28 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.34 0.34 0.28 0.09 0.09 0.09 0.28
0.00 0.17 0.17 0.17 0.00 0.00 0.17 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00
0.20 0.00 0.00 0.00 0.30 0.30 0.00 0.30 0.20 0.20 0.20 0.09 0.09 0.09 0.20
0.31 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.28 0.28 0.31 0.09 0.09 0.09 0.37
0.31 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.28 0.28 0.31 0.09 0.09 0.09 0.37
0.55 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.28 0.28 0.55 0.09 0.09 0.09 0.31
0.00 0.90 0.90 0.90 0.00 0.00 0.17 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00
0.40 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.28 0.28 0.40 0.09 0.09 0.09 0.31
0.28 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.76 0.76 0.28 0.09 0.09 0.09 0.28
0.09 0.00 0.00 0.00 0.09 0.09 0.00 0.09 0.09 0.09 0.09 0.86 0.86 0.86 0.09
0.20 0.00 0.00 0.00 0.21 0.21 0.00 0.21 0.20 0.20 0.20 0.09 0.09 0.09 0.20
0.25 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.25 0.25 0.25 0.09 0.09 0.09 0.25
0.28 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.34 0.34 0.28 0.09 0.09 0.09 0.28
0.76 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.28 0.28 0.55 0.09 0.09 0.09 0.31
0.20 0.00 0.00 0.00 0.43 0.43 0.00 0.34 0.20 0.20 0.20 0.09 0.09 0.09 0.20
0.31 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.28 0.28 0.31 0.09 0.09 0.09 0.37
0.31 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.28 0.28 0.31 0.09 0.09 0.09 0.37
0.31 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.28 0.28 0.31 0.09 0.09 0.09 0.37
0.00 0.90 0.90 0.90 0.00 0.00 0.17 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00
0.00 0.88 0.88 0.88 0.00 0.00 0.17 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00
0.31 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.28 0.28 0.31 0.09 0.09 0.09 0.38
0.31 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.28 0.28 0.31 0.09 0.09 0.09 0.37
0.28 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.68 0.68 0.28 0.09 0.09 0.09 0.28
0.55 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.28 0.28 0.69 0.09 0.09 0.09 0.31
0.36 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.28 0.28 0.36 0.09 0.09 0.09 0.31
0.00 0.92 0.92 0.92 0.00 0.00 0.17 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00
0.25 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.25 0.25 0.25 0.09 0.09 0.09 0.25
0.25 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.25 0.25 0.25 0.09 0.09 0.09 0.25
0.55 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.28 0.28 0.69 0.09 0.09 0.09 0.31
0.31 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.28 0.28 0.31 0.09 0.09 0.09 0.37
0.00 0.88 0.88 0.88 0.00 0.00 0.17 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00
0.55 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.28 0.28 0.65 0.09 0.09 0.09 0.31
0.28 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.34 0.34 0.28 0.09 0.09 0.09 0.28
0.20 0.00 0.00 0.00 0.34 0.34 0.00 0.78 0.20 0.20 0.20 0.09 0.09 0.09 0.20
0.40 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.28 0.28 0.40 0.09 0.09 0.09 0.31
0.31 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.28 0.28 0.31 0.09 0.09 0.09 0.37
0.31 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.28 0.28 0.31 0.09 0.09 0.09 0.80
0.36 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.28 0.28 0.36 0.09 0.09 0.09 0.31
0.31 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.28 0.28 0.31 0.09 0.09 0.09 0.37
0.22 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.22 0.22 0.22 0.09 0.09 0.09 0.22
0.40 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.28 0.28 0.40 0.09 0.09 0.09 0.31
0.25 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.25 0.25 0.25 0.09 0.09 0.09 0.25
0.25 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.25 0.25 0.25 0.09 0.09 0.09 0.25
0.20 0.00 0.00 0.00 0.30 0.30 0.00 0.30 0.20 0.20 0.20 0.09 0.09 0.09 0.20
0.09 0.00 0.00 0.00 0.09 0.09 0.00 0.09 0.09 0.09 0.09 0.64 0.64 0.64 0.09
0.00 0.88 0.88 0.88 0.00 0.00 0.17 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00
0.00 0.45 0.45 0.45 0.00 0.00 0.17 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00
0.00 0.69 0.69 0.69 0.00 0.00 0.17 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00
0.28 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.76 0.76 0.28 0.09 0.09 0.09 0.28
0.00 0.90 0.90 0.90 0.00 0.00 0.17 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00
0.55 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.28 0.28 0.55 0.09 0.09 0.09 0.31
0.31 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.28 0.28 0.31 0.09 0.09 0.09 0.37
0.00 0.90 0.90 0.90 0.00 0.00 0.17 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00
0.55 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.28 0.28 0.63 0.09 0.09 0.09 0.31
0.31 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.28 0.28 0.31 0.09 0.09 0.09 0.37
0.31 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.28 0.28 0.31 0.09 0.09 0.09 0.37
0.28 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.34 0.34 0.28 0.09 0.09 0.09 0.28
0.31 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.28 0.28 0.31 0.09 0.09 0.09 0.37
0.20 0.00 0.00 0.00 0.43 0.43 0.00 0.34 0.20 0.20 0.20 0.09 0.09 0.09 0.20
0.00 0.11 0.11 0.11 0.00 0.00 0.11 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00
0.55 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.28 0.28 0.69 0.09 0.09 0.09 0.31
0.40 0.00 0.00 0.00 0.20 0.20 0.00 0.20 0.28 0.28 0.40 0.09 0.09 0.09 0.31
0.09 0.00 0.00 0.00 0.09 0.09 0.00 0.09 0.09 0.09 0.09 0.68 0.68 0.68 0.09
0.11
0.00 0.00
0.00 0.00 0.84
0.00 0.00 0.20 0.20
0.00 0.00 0.20 0.20 0.31
0.00 0.00 0.20 0.20 0.31 0.55
0.00 0.00 0.20 0.20 0.37 0.31 0.31
0.88 0.11 0.00 0.00 0.00 0.00 0.00 0.00
0.45 0.11 0.00 0.00 0.00 0.00 0.00 0.00 0.45
0.00 0.00 0.20 0.20 0.38 0.31 0.31 0.37 0.00 0.00
0.91 0.11 0.00 0.00 0.00 0.00 0.00 0.00 0.88 0.45 0.00
0.00 0.00 0.20 0.20 0.31 0.40 0.40 0.31 0.00 0.00 0.31 0.00
0.00 0.00 0.26 0.26 0.20 0.20 0.20 0.20 0.00 0.00 0.20 0.00 0.20
0.45 0.11 0.00 0.00 0.00 0.00 0.00 0.00 0.45 0.50 0.00 0.45 0.00 0.00
0.45 0.11 0.00 0.00 0.00 0.00 0.00 0.00 0.45 0.50 0.00 0.45 0.00 0.00 0.63
0.45 0.11 0.00 0.00 0.00 0.00 0.00 0.00 0.45 0.50 0.00 0.45 0.00 0.00 0.63
0.00 0.00 0.20 0.20 0.25 0.25 0.25 0.25 0.00 0.00 0.25 0.00 0.25 0.20 0.00
0.00 0.00 0.30 0.30 0.20 0.20 0.20 0.20 0.00 0.00 0.20 0.00 0.20 0.26 0.00
0.00 0.00 0.20 0.20 0.31 0.55 0.77 0.31 0.00 0.00 0.31 0.00 0.40 0.20 0.00
0.00 0.00 0.20 0.20 0.37 0.31 0.31 0.48 0.00 0.00 0.37 0.00 0.31 0.20 0.00
0.00 0.00 0.34 0.34 0.20 0.20 0.20 0.20 0.00 0.00 0.20 0.00 0.20 0.26 0.00
0.00 0.00 0.09 0.09 0.09 0.09 0.09 0.09 0.00 0.00 0.09 0.00 0.09 0.09 0.00
0.00 0.00 0.20 0.20 0.28 0.28 0.28 0.28 0.00 0.00 0.28 0.00 0.28 0.20 0.00
0.00 0.00 0.20 0.20 0.28 0.28 0.28 0.28 0.00 0.00 0.28 0.00 0.28 0.20 0.00
0.00 0.00 0.09 0.09 0.09 0.09 0.09 0.09 0.00 0.00 0.09 0.00 0.09 0.09 0.00
0.00 0.00 0.20 0.20 0.28 0.28 0.28 0.28 0.00 0.00 0.28 0.00 0.28 0.20 0.00
0.17 0.11 0.00 0.00 0.00 0.00 0.00 0.00 0.17 0.17 0.00 0.17 0.00 0.00 0.17
0.00 0.00 0.30 0.30 0.20 0.20 0.20 0.20 0.00 0.00 0.20 0.00 0.20 0.26 0.00
0.00 0.00 0.20 0.20 0.37 0.31 0.31 0.84 0.00 0.00 0.37 0.00 0.31 0.20 0.00
0.00 0.00 0.20 0.20 0.37 0.31 0.31 0.84 0.00 0.00 0.37 0.00 0.31 0.20 0.00
0.00 0.00 0.20 0.20 0.31 0.55 0.85 0.31 0.00 0.00 0.31 0.00 0.40 0.20 0.00
0.91 0.11 0.00 0.00 0.00 0.00 0.00 0.00 0.88 0.45 0.00 0.91 0.00 0.00 0.45
0.00 0.00 0.20 0.20 0.31 0.40 0.40 0.31 0.00 0.00 0.31 0.00 0.78 0.20 0.00
0.00 0.00 0.20 0.20 0.28 0.28 0.28 0.28 0.00 0.00 0.28 0.00 0.28 0.20 0.00
0.00 0.00 0.09 0.09 0.09 0.09 0.09 0.09 0.00 0.00 0.09 0.00 0.09 0.09 0.00
0.00 0.00 0.21 0.21 0.20 0.20 0.20 0.20 0.00 0.00 0.20 0.00 0.20 0.21 0.00
0.00 0.00 0.20 0.20 0.25 0.25 0.25 0.25 0.00 0.00 0.25 0.00 0.25 0.20 0.00
0.00 0.00 0.20 0.20 0.28 0.28 0.28 0.28 0.00 0.00 0.28 0.00 0.28 0.20 0.00
0.00 0.00 0.20 0.20 0.31 0.82 0.55 0.31 0.00 0.00 0.31 0.00 0.40 0.20 0.00
0.00 0.00 0.80 0.80 0.20 0.20 0.20 0.20 0.00 0.00 0.20 0.00 0.20 0.26 0.00
0.00 0.00 0.20 0.20 0.37 0.31 0.31 0.84 0.00 0.00 0.37 0.00 0.31 0.20 0.00
0.00 0.00 0.20 0.20 0.37 0.31 0.31 0.83 0.00 0.00 0.37 0.00 0.31 0.20 0.00
0.00 0.00 0.20 0.20 0.37 0.31 0.31 0.84 0.00 0.00 0.37 0.00 0.31 0.20 0.00
0.91 0.11 0.00 0.00 0.00 0.00 0.00 0.00 0.88 0.45 0.00 0.91 0.00 0.00 0.45
0.88 0.11 0.00 0.00 0.00 0.00 0.00 0.00 0.88 0.45 0.00 0.88 0.00 0.00 0.45
0.00 0.00 0.20 0.20 0.38 0.31 0.31 0.37 0.00 0.00 0.70 0.00 0.31 0.20 0.00
0.00 0.00 0.20 0.20 0.37 0.31 0.31 0.84 0.00 0.00 0.37 0.00 0.31 0.20 0.00
0.00 0.00 0.20 0.20 0.28 0.28 0.28 0.28 0.00 0.00 0.28 0.00 0.28 0.20 0.00
0.00 0.00 0.20 0.20 0.31 0.55 0.55 0.31 0.00 0.00 0.31 0.00 0.40 0.20 0.00
0.00 0.00 0.20 0.20 0.31 0.36 0.36 0.31 0.00 0.00 0.31 0.00 0.36 0.20 0.00
0.90 0.11 0.00 0.00 0.00 0.00 0.00 0.00 0.88 0.45 0.00 0.90 0.00 0.00 0.45
0.00 0.00 0.20 0.20 0.25 0.25 0.25 0.25 0.00 0.00 0.25 0.00 0.25 0.20 0.00
0.00 0.00 0.20 0.20 0.25 0.25 0.25 0.25 0.00 0.00 0.25 0.00 0.25 0.20 0.00
0.00 0.00 0.20 0.20 0.31 0.55 0.55 0.31 0.00 0.00 0.31 0.00 0.40 0.20 0.00
0.00 0.00 0.20 0.20 0.37 0.31 0.31 0.81 0.00 0.00 0.37 0.00 0.31 0.20 0.00
0.88 0.11 0.00 0.00 0.00 0.00 0.00 0.00 0.94 0.45 0.00 0.88 0.00 0.00 0.45
0.00 0.00 0.20 0.20 0.31 0.55 0.55 0.31 0.00 0.00 0.31 0.00 0.40 0.20 0.00
0.00 0.00 0.20 0.20 0.28 0.28 0.28 0.28 0.00 0.00 0.28 0.00 0.28 0.20 0.00
0.00 0.00 0.34 0.34 0.20 0.20 0.20 0.20 0.00 0.00 0.20 0.00 0.20 0.26 0.00
0.00 0.00 0.20 0.20 0.31 0.40 0.40 0.31 0.00 0.00 0.31 0.00 0.86 0.20 0.00
0.00 0.00 0.20 0.20 0.37 0.31 0.31 0.81 0.00 0.00 0.37 0.00 0.31 0.20 0.00
0.00 0.00 0.20 0.20 0.59 0.31 0.31 0.37 0.00 0.00 0.38 0.00 0.31 0.20 0.00
0.00 0.00 0.20 0.20 0.31 0.36 0.36 0.31 0.00 0.00 0.31 0.00 0.36 0.20 0.00
0.00 0.00 0.20 0.20 0.37 0.31 0.31 0.84 0.00 0.00 0.37 0.00 0.31 0.20 0.00
0.00 0.00 0.20 0.20 0.22 0.22 0.22 0.22 0.00 0.00 0.22 0.00 0.22 0.20 0.00
0.00 0.00 0.20 0.20 0.31 0.40 0.40 0.31 0.00 0.00 0.31 0.00 0.95 0.20 0.00
0.00 0.00 0.20 0.20 0.25 0.25 0.25 0.25 0.00 0.00 0.25 0.00 0.25 0.20 0.00
0.00 0.00 0.20 0.20 0.25 0.25 0.25 0.25 0.00 0.00 0.25 0.00 0.25 0.20 0.00
0.00 0.00 0.30 0.30 0.20 0.20 0.20 0.20 0.00 0.00 0.20 0.00 0.20 0.26 0.00
0.00 0.00 0.09 0.09 0.09 0.09 0.09 0.09 0.00 0.00 0.09 0.00 0.09 0.09 0.00
0.88 0.11 0.00 0.00 0.00 0.00 0.00 0.00 0.93 0.45 0.00 0.88 0.00 0.00 0.45
0.45 0.11 0.00 0.00 0.00 0.00 0.00 0.00 0.45 0.50 0.00 0.45 0.00 0.00 0.63
0.69 0.11 0.00 0.00 0.00 0.00 0.00 0.00 0.69 0.45 0.00 0.69 0.00 0.00 0.45
0.00 0.00 0.20 0.20 0.28 0.28 0.28 0.28 0.00 0.00 0.28 0.00 0.28 0.20 0.00
0.91 0.11 0.00 0.00 0.00 0.00 0.00 0.00 0.88 0.45 0.00 0.91 0.00 0.00 0.45
0.00 0.00 0.20 0.20 0.31 0.55 0.83 0.31 0.00 0.00 0.31 0.00 0.40 0.20 0.00
0.00 0.00 0.20 0.20 0.37 0.31 0.31 0.81 0.00 0.00 0.37 0.00 0.31 0.20 0.00
0.91 0.11 0.00 0.00 0.00 0.00 0.00 0.00 0.88 0.45 0.00 0.91 0.00 0.00 0.45
0.00 0.00 0.20 0.20 0.31 0.55 0.55 0.31 0.00 0.00 0.31 0.00 0.40 0.20 0.00
0.00 0.00 0.20 0.20 0.37 0.31 0.31 0.83 0.00 0.00 0.37 0.00 0.31 0.20 0.00
0.00 0.00 0.20 0.20 0.37 0.31 0.31 0.81 0.00 0.00 0.37 0.00 0.31 0.20 0.00
0.00 0.00 0.20 0.20 0.28 0.28 0.28 0.28 0.00 0.00 0.28 0.00 0.28 0.20 0.00
0.00 0.00 0.20 0.20 0.37 0.31 0.31 0.81 0.00 0.00 0.37 0.00 0.31 0.20 0.00
0.00 0.00 0.74 0.74 0.20 0.20 0.20 0.20 0.00 0.00 0.20 0.00 0.20 0.26 0.00
0.11 0.39 0.00 0.00 0.00 0.00 0.00 0.00 0.11 0.11 0.00 0.11 0.00 0.00 0.11
0.00 0.00 0.20 0.20 0.31 0.55 0.55 0.31 0.00 0.00 0.31 0.00 0.40 0.20 0.00
0.00 0.00 0.20 0.20 0.31 0.40 0.40 0.31 0.00 0.00 0.31 0.00 0.53 0.20 0.00
0.00 0.00 0.09 0.09 0.09 0.09 0.09 0.09 0.00 0.00 0.09 0.00 0.09 0.09 0.00
0.75
0.00 0.00
0.00 0.00 0.20
0.00 0.00 0.25 0.20
0.00 0.00 0.25 0.20 0.31
0.00 0.00 0.20 0.30 0.20 0.20
0.00 0.00 0.09 0.09 0.09 0.09 0.09
0.00 0.00 0.25 0.20 0.28 0.28 0.20 0.09
0.00 0.00 0.25 0.20 0.28 0.28 0.20 0.09 0.34
0.00 0.00 0.09 0.09 0.09 0.09 0.09 0.54 0.09 0.09
0.00 0.00 0.25 0.20 0.28 0.28 0.20 0.09 0.51 0.34 0.09
0.17 0.17 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00
0.00 0.00 0.20 0.31 0.20 0.20 0.30 0.09 0.20 0.20 0.09 0.20 0.00
0.00 0.00 0.25 0.20 0.31 0.48 0.20 0.09 0.28 0.28 0.09 0.28 0.00 0.20
0.00 0.00 0.25 0.20 0.31 0.48 0.20 0.09 0.28 0.28 0.09 0.28 0.00 0.20 0.96
0.00 0.00 0.25 0.20 0.77 0.31 0.20 0.09 0.28 0.28 0.09 0.28 0.00 0.20 0.31
0.45 0.45 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.17 0.00 0.00
0.00 0.00 0.25 0.20 0.40 0.31 0.20 0.09 0.28 0.28 0.09 0.28 0.00 0.20 0.31
0.00 0.00 0.25 0.20 0.28 0.28 0.20 0.09 0.34 0.75 0.09 0.34 0.00 0.20 0.28
0.00 0.00 0.09 0.09 0.09 0.09 0.09 0.86 0.09 0.09 0.54 0.09 0.00 0.09 0.09
0.00 0.00 0.20 0.21 0.20 0.20 0.21 0.09 0.20 0.20 0.09 0.20 0.00 0.21 0.20
0.00 0.00 0.78 0.20 0.25 0.25 0.20 0.09 0.25 0.25 0.09 0.25 0.00 0.20 0.25
0.00 0.00 0.25 0.20 0.28 0.28 0.20 0.09 0.51 0.34 0.09 0.86 0.00 0.20 0.28
0.00 0.00 0.25 0.20 0.55 0.31 0.20 0.09 0.28 0.28 0.09 0.28 0.00 0.20 0.31
0.00 0.00 0.20 0.30 0.20 0.20 0.34 0.09 0.20 0.20 0.09 0.20 0.00 0.30 0.20
0.00 0.00 0.25 0.20 0.31 0.48 0.20 0.09 0.28 0.28 0.09 0.28 0.00 0.20 0.96
0.00 0.00 0.25 0.20 0.31 0.48 0.20 0.09 0.28 0.28 0.09 0.28 0.00 0.20 0.83
0.00 0.00 0.25 0.20 0.31 0.48 0.20 0.09 0.28 0.28 0.09 0.28 0.00 0.20 0.86
0.45 0.45 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.17 0.00 0.00
0.45 0.45 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.17 0.00 0.00
0.00 0.00 0.25 0.20 0.31 0.37 0.20 0.09 0.28 0.28 0.09 0.28 0.00 0.20 0.37
0.00 0.00 0.25 0.20 0.31 0.48 0.20 0.09 0.28 0.28 0.09 0.28 0.00 0.20 0.89
0.00 0.00 0.25 0.20 0.28 0.28 0.20 0.09 0.34 0.68 0.09 0.34 0.00 0.20 0.28
0.00 0.00 0.25 0.20 0.55 0.31 0.20 0.09 0.28 0.28 0.09 0.28 0.00 0.20 0.31
0.00 0.00 0.25 0.20 0.36 0.31 0.20 0.09 0.28 0.28 0.09 0.28 0.00 0.20 0.31
0.45 0.45 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.17 0.00 0.00
0.00 0.00 0.43 0.20 0.25 0.25 0.20 0.09 0.25 0.25 0.09 0.25 0.00 0.20 0.25
0.00 0.00 0.78 0.20 0.25 0.25 0.20 0.09 0.25 0.25 0.09 0.25 0.00 0.20 0.25
0.00 0.00 0.25 0.20 0.55 0.31 0.20 0.09 0.28 0.28 0.09 0.28 0.00 0.20 0.31
0.00 0.00 0.25 0.20 0.31 0.48 0.20 0.09 0.28 0.28 0.09 0.28 0.00 0.20 0.81
0.45 0.45 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.17 0.00 0.00
0.00 0.00 0.25 0.20 0.55 0.31 0.20 0.09 0.28 0.28 0.09 0.28 0.00 0.20 0.31
0.00 0.00 0.25 0.20 0.28 0.28 0.20 0.09 0.52 0.34 0.09 0.51 0.00 0.20 0.28
0.00 0.00 0.20 0.30 0.20 0.20 0.78 0.09 0.20 0.20 0.09 0.20 0.00 0.30 0.20
0.00 0.00 0.25 0.20 0.40 0.31 0.20 0.09 0.28 0.28 0.09 0.28 0.00 0.20 0.31
0.00 0.00 0.25 0.20 0.31 0.48 0.20 0.09 0.28 0.28 0.09 0.28 0.00 0.20 0.81
0.00 0.00 0.25 0.20 0.31 0.37 0.20 0.09 0.28 0.28 0.09 0.28 0.00 0.20 0.37
0.00 0.00 0.25 0.20 0.36 0.31 0.20 0.09 0.28 0.28 0.09 0.28 0.00 0.20 0.31
0.00 0.00 0.25 0.20 0.31 0.48 0.20 0.09 0.28 0.28 0.09 0.28 0.00 0.20 0.86
0.00 0.00 0.22 0.20 0.22 0.22 0.20 0.09 0.22 0.22 0.09 0.22 0.00 0.20 0.22
0.00 0.00 0.25 0.20 0.40 0.31 0.20 0.09 0.28 0.28 0.09 0.28 0.00 0.20 0.31
0.00 0.00 0.78 0.20 0.25 0.25 0.20 0.09 0.25 0.25 0.09 0.25 0.00 0.20 0.25
0.00 0.00 0.78 0.20 0.25 0.25 0.20 0.09 0.25 0.25 0.09 0.25 0.00 0.20 0.25
0.00 0.00 0.20 0.31 0.20 0.20 0.30 0.09 0.20 0.20 0.09 0.20 0.00 0.40 0.20
0.00 0.00 0.09 0.09 0.09 0.09 0.09 0.64 0.09 0.09 0.54 0.09 0.00 0.09 0.09
0.45 0.45 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.17 0.00 0.00
0.75 0.75 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.17 0.00 0.00
0.45 0.45 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.17 0.00 0.00
0.00 0.00 0.25 0.20 0.28 0.28 0.20 0.09 0.34 0.75 0.09 0.34 0.00 0.20 0.28
0.45 0.45 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.17 0.00 0.00
0.00 0.00 0.25 0.20 0.77 0.31 0.20 0.09 0.28 0.28 0.09 0.28 0.00 0.20 0.31
0.00 0.00 0.25 0.20 0.31 0.48 0.20 0.09 0.28 0.28 0.09 0.28 0.00 0.20 0.81
0.45 0.45 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.17 0.00 0.00
0.00 0.00 0.25 0.20 0.55 0.31 0.20 0.09 0.28 0.28 0.09 0.28 0.00 0.20 0.31
0.00 0.00 0.25 0.20 0.31 0.48 0.20 0.09 0.28 0.28 0.09 0.28 0.00 0.20 0.83
0.00 0.00 0.25 0.20 0.31 0.48 0.20 0.09 0.28 0.28 0.09 0.28 0.00 0.20 0.81
0.00 0.00 0.25 0.20 0.28 0.28 0.20 0.09 0.46 0.34 0.09 0.46 0.00 0.20 0.28
0.00 0.00 0.25 0.20 0.31 0.48 0.20 0.09 0.28 0.28 0.09 0.28 0.00 0.20 0.81
0.00 0.00 0.20 0.30 0.20 0.20 0.34 0.09 0.20 0.20 0.09 0.20 0.00 0.30 0.20
0.11 0.11 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.11 0.00 0.00
0.00 0.00 0.25 0.20 0.55 0.31 0.20 0.09 0.28 0.28 0.09 0.28 0.00 0.20 0.31
0.00 0.00 0.25 0.20 0.40 0.31 0.20 0.09 0.28 0.28 0.09 0.28 0.00 0.20 0.31
0.00 0.00 0.09 0.09 0.09 0.09 0.09 0.68 0.09 0.09 0.54 0.09 0.00 0.09 0.09
0.31
0.00 0.00
0.31 0.40 0.00
0.28 0.28 0.00 0.28
0.09 0.09 0.00 0.09 0.09
0.20 0.20 0.00 0.20 0.20 0.09
0.25 0.25 0.00 0.25 0.25 0.09 0.20
0.28 0.28 0.00 0.28 0.34 0.09 0.20 0.25
0.31 0.55 0.00 0.40 0.28 0.09 0.20 0.25 0.28
0.20 0.20 0.00 0.20 0.20 0.09 0.21 0.20 0.20 0.20
0.98 0.31 0.00 0.31 0.28 0.09 0.20 0.25 0.28 0.31 0.20
0.83 0.31 0.00 0.31 0.28 0.09 0.20 0.25 0.28 0.31 0.20 0.83
0.86 0.31 0.00 0.31 0.28 0.09 0.20 0.25 0.28 0.31 0.20 0.86 0.83
0.00 0.00 0.94 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00
0.00 0.00 0.88 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.88
0.37 0.31 0.00 0.31 0.28 0.09 0.20 0.25 0.28 0.31 0.20 0.37 0.37 0.37 0.00
0.89 0.31 0.00 0.31 0.28 0.09 0.20 0.25 0.28 0.31 0.20 0.89 0.83 0.86 0.00
0.28 0.28 0.00 0.28 0.68 0.09 0.20 0.25 0.34 0.28 0.20 0.28 0.28 0.28 0.00
0.31 0.55 0.00 0.40 0.28 0.09 0.20 0.25 0.28 0.55 0.20 0.31 0.31 0.31 0.00
0.31 0.36 0.00 0.36 0.28 0.09 0.20 0.25 0.28 0.36 0.20 0.31 0.31 0.31 0.00
0.00 0.00 0.90 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.90
0.25 0.25 0.00 0.25 0.25 0.09 0.20 0.43 0.25 0.25 0.20 0.25 0.25 0.25 0.00
0.25 0.25 0.00 0.25 0.25 0.09 0.20 0.79 0.25 0.25 0.20 0.25 0.25 0.25 0.00
0.31 0.55 0.00 0.40 0.28 0.09 0.20 0.25 0.28 0.55 0.20 0.31 0.31 0.31 0.00
0.81 0.31 0.00 0.31 0.28 0.09 0.20 0.25 0.28 0.31 0.20 0.81 0.81 0.81 0.00
0.00 0.00 0.88 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.88
0.31 0.55 0.00 0.40 0.28 0.09 0.20 0.25 0.28 0.55 0.20 0.31 0.31 0.31 0.00
0.28 0.28 0.00 0.28 0.34 0.09 0.20 0.25 0.51 0.28 0.20 0.28 0.28 0.28 0.00
0.20 0.20 0.00 0.20 0.20 0.09 0.21 0.20 0.20 0.20 0.34 0.20 0.20 0.20 0.00
0.31 0.40 0.00 0.78 0.28 0.09 0.20 0.25 0.28 0.40 0.20 0.31 0.31 0.31 0.00
0.81 0.31 0.00 0.31 0.28 0.09 0.20 0.25 0.28 0.31 0.20 0.81 0.81 0.81 0.00
0.37 0.31 0.00 0.31 0.28 0.09 0.20 0.25 0.28 0.31 0.20 0.37 0.37 0.37 0.00
0.31 0.36 0.00 0.36 0.28 0.09 0.20 0.25 0.28 0.36 0.20 0.31 0.31 0.31 0.00
0.86 0.31 0.00 0.31 0.28 0.09 0.20 0.25 0.28 0.31 0.20 0.86 0.83 0.91 0.00
0.22 0.22 0.00 0.22 0.22 0.09 0.20 0.22 0.22 0.22 0.20 0.22 0.22 0.22 0.00
0.31 0.40 0.00 0.78 0.28 0.09 0.20 0.25 0.28 0.40 0.20 0.31 0.31 0.31 0.00
0.25 0.25 0.00 0.25 0.25 0.09 0.20 0.79 0.25 0.25 0.20 0.25 0.25 0.25 0.00
0.25 0.25 0.00 0.25 0.25 0.09 0.20 0.79 0.25 0.25 0.20 0.25 0.25 0.25 0.00
0.20 0.20 0.00 0.20 0.20 0.09 0.21 0.20 0.20 0.20 0.30 0.20 0.20 0.20 0.00
0.09 0.09 0.00 0.09 0.09 0.64 0.09 0.09 0.09 0.09 0.09 0.09 0.09 0.09 0.00
0.00 0.00 0.88 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.88
0.00 0.00 0.45 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.45
0.00 0.00 0.69 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.69
0.28 0.28 0.00 0.28 0.83 0.09 0.20 0.25 0.34 0.28 0.20 0.28 0.28 0.28 0.00
0.00 0.00 0.94 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.96
0.31 0.83 0.00 0.40 0.28 0.09 0.20 0.25 0.28 0.55 0.20 0.31 0.31 0.31 0.00
0.81 0.31 0.00 0.31 0.28 0.09 0.20 0.25 0.28 0.31 0.20 0.81 0.81 0.81 0.00
0.00 0.00 0.91 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.91
0.31 0.55 0.00 0.40 0.28 0.09 0.20 0.25 0.28 0.55 0.20 0.31 0.31 0.31 0.00
0.83 0.31 0.00 0.31 0.28 0.09 0.20 0.25 0.28 0.31 0.20 0.83 0.94 0.83 0.00
0.81 0.31 0.00 0.31 0.28 0.09 0.20 0.25 0.28 0.31 0.20 0.81 0.81 0.81 0.00
0.28 0.28 0.00 0.28 0.34 0.09 0.20 0.25 0.46 0.28 0.20 0.28 0.28 0.28 0.00
0.81 0.31 0.00 0.31 0.28 0.09 0.20 0.25 0.28 0.31 0.20 0.81 0.81 0.81 0.00
0.20 0.20 0.00 0.20 0.20 0.09 0.21 0.20 0.20 0.20 0.74 0.20 0.20 0.20 0.00
0.00 0.00 0.11 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.11
0.31 0.55 0.00 0.40 0.28 0.09 0.20 0.25 0.28 0.55 0.20 0.31 0.31 0.31 0.00
0.31 0.40 0.00 0.53 0.28 0.09 0.20 0.25 0.28 0.40 0.20 0.31 0.31 0.31 0.00
0.09 0.09 0.00 0.09 0.09 0.68 0.09 0.09 0.09 0.09 0.09 0.09 0.09 0.09 0.00
0.00
0.00 0.37
0.00 0.28 0.28
0.00 0.31 0.31 0.28
0.00 0.31 0.31 0.28 0.36
0.88 0.00 0.00 0.00 0.00 0.00
0.00 0.25 0.25 0.25 0.25 0.25 0.00
0.00 0.25 0.25 0.25 0.25 0.25 0.00 0.43
0.00 0.31 0.31 0.28 0.85 0.36 0.00 0.25 0.25
0.00 0.37 0.81 0.28 0.31 0.31 0.00 0.25 0.25 0.31
0.88 0.00 0.00 0.00 0.00 0.00 0.88 0.00 0.00 0.00 0.00
0.00 0.31 0.31 0.28 0.65 0.36 0.00 0.25 0.25 0.65 0.31 0.00
0.00 0.28 0.28 0.34 0.28 0.28 0.00 0.25 0.25 0.28 0.28 0.00 0.28
0.00 0.20 0.20 0.20 0.20 0.20 0.00 0.20 0.20 0.20 0.20 0.00 0.20 0.20
0.00 0.31 0.31 0.28 0.40 0.36 0.00 0.25 0.25 0.40 0.31 0.00 0.40 0.28 0.20
0.00 0.37 0.81 0.28 0.31 0.31 0.00 0.25 0.25 0.31 0.86 0.00 0.31 0.28 0.20
0.00 0.38 0.37 0.28 0.31 0.31 0.00 0.25 0.25 0.31 0.37 0.00 0.31 0.28 0.20
0.00 0.31 0.31 0.28 0.36 0.91 0.00 0.25 0.25 0.36 0.31 0.00 0.36 0.28 0.20
0.00 0.37 0.86 0.28 0.31 0.31 0.00 0.25 0.25 0.31 0.81 0.00 0.31 0.28 0.20
0.00 0.22 0.22 0.22 0.22 0.22 0.00 0.22 0.22 0.22 0.22 0.00 0.22 0.22 0.20
0.00 0.31 0.31 0.28 0.40 0.36 0.00 0.25 0.25 0.40 0.31 0.00 0.40 0.28 0.20
0.00 0.25 0.25 0.25 0.25 0.25 0.00 0.43 0.79 0.25 0.25 0.00 0.25 0.25 0.20
0.00 0.25 0.25 0.25 0.25 0.25 0.00 0.43 0.80 0.25 0.25 0.00 0.25 0.25 0.20
0.00 0.20 0.20 0.20 0.20 0.20 0.00 0.20 0.20 0.20 0.20 0.00 0.20 0.20 0.30
0.00 0.09 0.09 0.09 0.09 0.09 0.00 0.09 0.09 0.09 0.09 0.00 0.09 0.09 0.09
0.88 0.00 0.00 0.00 0.00 0.00 0.88 0.00 0.00 0.00 0.00 0.93 0.00 0.00 0.00
0.45 0.00 0.00 0.00 0.00 0.00 0.45 0.00 0.00 0.00 0.00 0.45 0.00 0.00 0.00
0.69 0.00 0.00 0.00 0.00 0.00 0.69 0.00 0.00 0.00 0.00 0.69 0.00 0.00 0.00
0.00 0.28 0.28 0.68 0.28 0.28 0.00 0.25 0.25 0.28 0.28 0.00 0.28 0.34 0.20
0.88 0.00 0.00 0.00 0.00 0.00 0.90 0.00 0.00 0.00 0.00 0.88 0.00 0.00 0.00
0.00 0.31 0.31 0.28 0.55 0.36 0.00 0.25 0.25 0.55 0.31 0.00 0.55 0.28 0.20
0.00 0.37 0.81 0.28 0.31 0.31 0.00 0.25 0.25 0.31 0.88 0.00 0.31 0.28 0.20
0.88 0.00 0.00 0.00 0.00 0.00 0.90 0.00 0.00 0.00 0.00 0.88 0.00 0.00 0.00
0.00 0.31 0.31 0.28 0.63 0.36 0.00 0.25 0.25 0.63 0.31 0.00 0.63 0.28 0.20
0.00 0.37 0.83 0.28 0.31 0.31 0.00 0.25 0.25 0.31 0.81 0.00 0.31 0.28 0.20
0.00 0.37 0.81 0.28 0.31 0.31 0.00 0.25 0.25 0.31 0.88 0.00 0.31 0.28 0.20
0.00 0.28 0.28 0.34 0.28 0.28 0.00 0.25 0.25 0.28 0.28 0.00 0.28 0.46 0.20
0.00 0.37 0.81 0.28 0.31 0.31 0.00 0.25 0.25 0.31 0.88 0.00 0.31 0.28 0.20
0.00 0.20 0.20 0.20 0.20 0.20 0.00 0.20 0.20 0.20 0.20 0.00 0.20 0.20 0.34
0.11 0.00 0.00 0.00 0.00 0.00 0.11 0.00 0.00 0.00 0.00 0.11 0.00 0.00 0.00
0.00 0.31 0.31 0.28 0.87 0.36 0.00 0.25 0.25 0.85 0.31 0.00 0.65 0.28 0.20
0.00 0.31 0.31 0.28 0.40 0.36 0.00 0.25 0.25 0.40 0.31 0.00 0.40 0.28 0.20
0.00 0.09 0.09 0.09 0.09 0.09 0.00 0.09 0.09 0.09 0.09 0.00 0.09 0.09 0.09
0.31
0.31 0.37
0.36 0.31 0.31
0.31 0.81 0.37 0.31
0.22 0.22 0.22 0.22 0.22
0.86 0.31 0.31 0.36 0.31 0.22
0.25 0.25 0.25 0.25 0.25 0.22 0.25
0.25 0.25 0.25 0.25 0.25 0.22 0.25 0.79
0.20 0.20 0.20 0.20 0.20 0.20 0.20 0.20 0.20
0.09 0.09 0.09 0.09 0.09 0.09 0.09 0.09 0.09 0.09
0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00
0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.45
0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.69 0.45
0.28 0.28 0.28 0.28 0.28 0.22 0.28 0.25 0.25 0.20 0.09 0.00 0.00 0.00
0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.88 0.45 0.69 0.00
0.40 0.31 0.31 0.36 0.31 0.22 0.40 0.25 0.25 0.20 0.09 0.00 0.00 0.00 0.28
0.31 0.86 0.37 0.31 0.81 0.22 0.31 0.25 0.25 0.20 0.09 0.00 0.00 0.00 0.28
0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.88 0.45 0.69 0.00
0.40 0.31 0.31 0.36 0.31 0.22 0.40 0.25 0.25 0.20 0.09 0.00 0.00 0.00 0.28
0.31 0.81 0.37 0.31 0.83 0.22 0.31 0.25 0.25 0.20 0.09 0.00 0.00 0.00 0.28
0.31 0.86 0.37 0.31 0.81 0.22 0.31 0.25 0.25 0.20 0.09 0.00 0.00 0.00 0.28
0.28 0.28 0.28 0.28 0.28 0.22 0.28 0.25 0.25 0.20 0.09 0.00 0.00 0.00 0.34
0.31 0.86 0.37 0.31 0.81 0.22 0.31 0.25 0.25 0.20 0.09 0.00 0.00 0.00 0.28
0.20 0.20 0.20 0.20 0.20 0.20 0.20 0.20 0.20 0.30 0.09 0.00 0.00 0.00 0.20
0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.11 0.11 0.11 0.00
0.40 0.31 0.31 0.36 0.31 0.22 0.40 0.25 0.25 0.20 0.09 0.00 0.00 0.00 0.28
0.53 0.31 0.31 0.36 0.31 0.22 0.53 0.25 0.25 0.20 0.09 0.00 0.00 0.00 0.28
0.09 0.09 0.09 0.09 0.09 0.09 0.09 0.09 0.09 0.09 0.64 0.00 0.00 0.00 0.09
0.00
0.00 0.31
0.91 0.00 0.00
0.00 0.55 0.31 0.00
0.00 0.31 0.81 0.00 0.31
0.00 0.31 0.97 0.00 0.31 0.81
0.00 0.28 0.28 0.00 0.28 0.28 0.28
0.00 0.31 0.94 0.00 0.31 0.81 0.94 0.28
0.00 0.20 0.20 0.00 0.20 0.20 0.20 0.20 0.20
0.11 0.00 0.00 0.11 0.00 0.00 0.00 0.00 0.00 0.00
0.00 0.55 0.31 0.00 0.63 0.31 0.31 0.28 0.31 0.20 0.00
0.00 0.40 0.31 0.00 0.40 0.31 0.31 0.28 0.31 0.20 0.00 0.40
0.00 0.09 0.09 0.00 0.09 0.09 0.09 0.09 0.09 0.09 0.00 0.09 0.09
Number of obs: 1729, groups: Site, 17; Species, 152; dummy, 1
Conditional model:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 1.6633 1.3864 1.200 0.23025
sSeedMass -0.3205 0.1166 -2.748 0.00599 **
sSeedShape -0.3606 0.1331 -2.709 0.00674 **
sSeedN -0.2142 0.1144 -1.872 0.06118 .
sAltitude -0.6441 0.3159 -2.039 0.04143 *
sHum 0.1031 0.3041 0.339 0.73456
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Dispersion model:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 0.519708 0.018722 27.759 < 2e-16 ***
sAltitude -0.120838 0.024140 -5.006 5.57e-07 ***
sHum -0.009928 0.026301 -0.377 0.706
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
simulateResiduals (model5, plot = TRUE ) # Unconditional simulations
Object of Class DHARMa with simulated residuals based on 250 simulations with refit = FALSE . See ?DHARMa::simulateResiduals for help.
Scaled residual values: 0.836 0.808 0.916 0.956 0.848 0.852 0.252 0.296 0.264 0.256 0.248 0.264 0.24 0.244 0.232 0.184 0.216 0.288 0.796 0.796 ...
Prepare+scale data:
= as.data.frame (EcoData:: seedBank)$ sAltitude = scale (data$ Altitude)$ sSeedMass = scale (data$ SeedMass)$ sSeedShape = scale (data$ SeedShape)$ sSeedN = scale (data$ SeedN)$ sSeedPr = scale (data$ SeedPr)$ sDormRank = scale (data$ DormRank)$ sTemp = scale (data$ Temp)$ sHum = scale (data$ Humidity)$ sNitro = scale (data$ Nitrogen)$ sGrazing = scale (data$ Grazing)$ sMGT = scale (data$ MGT)$ sJwidth = scale (data$ Jwidth)$ sEpiStein = scale (data$ EpiStein)$ sMGR = scale (data$ MGR)$ sT95 = scale (data$ T95)# Let's remove NAs beforehand: = rownames (model.matrix (SBDensity~ sAltitude + sSeedMass + sSeedShape + sSeedN + + sDormRank + sTemp + sHum + sNitro + sMGT + + sEpiStein + sT95 + + sGrazing + Site + Species, data = data))= data[rows, ]
= glmer (SBPA~ + sSeedShape + sSeedN + + sHum + 1 | Site) + (sAltitude| Species),data = data, family = binomial ())
Warning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
Model failed to converge with max|grad| = 0.0112646 (tol = 0.002, component 1)
Generalized linear mixed model fit by maximum likelihood (Laplace
Approximation) [glmerMod]
Family: binomial ( logit )
Formula: SBPA ~ sSeedMass + sSeedShape + sSeedN + sAltitude + sHum + (1 |
Site) + (sAltitude | Species)
Data: data
AIC BIC logLik -2*log(L) df.resid
1368.9 1423.4 -674.4 1348.9 1719
Scaled residuals:
Min 1Q Median 3Q Max
-2.5936 -0.2658 -0.1017 0.2001 3.3351
Random effects:
Groups Name Variance Std.Dev. Corr
Species (Intercept) 8.16403 2.8573
sAltitude 4.39310 2.0960 -0.13
Site (Intercept) 0.06797 0.2607
Number of obs: 1729, groups: Species, 152; Site, 17
Fixed effects:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -2.6286 0.4066 -6.465 1.01e-10 ***
sSeedMass -1.3671 0.6432 -2.125 0.0335 *
sSeedShape -0.5807 0.3099 -1.873 0.0610 .
sSeedN -2.1354 1.3164 -1.622 0.1048
sAltitude -1.3931 0.3365 -4.140 3.47e-05 ***
sHum -0.1021 0.1308 -0.781 0.4350
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Correlation of Fixed Effects:
(Intr) sSdMss sSdShp sSeedN sAlttd
sSeedMass 0.221
sSeedShape 0.033 0.062
sSeedN 0.336 0.060 -0.066
sAltitude 0.132 0.142 0.079 0.012
sHum 0.023 0.002 -0.003 -0.011 0.229
optimizer (Nelder_Mead) convergence code: 0 (OK)
Model failed to converge with max|grad| = 0.0112646 (tol = 0.002, component 1)
Model did not converge, but there is a trick which often helps. The default optimizer in lme4 is not the best optimizer, changing it to ‘bobyqa’ often helps with convergence issues
= glmer (SBPA~ + sSeedShape + sSeedN + + sHum + 1 | Site) + (sAltitude| Species),data = data, family = binomial (),control = glmerControl ('bobyqa' ))summary (model1)
Generalized linear mixed model fit by maximum likelihood (Laplace
Approximation) [glmerMod]
Family: binomial ( logit )
Formula: SBPA ~ sSeedMass + sSeedShape + sSeedN + sAltitude + sHum + (1 |
Site) + (sAltitude | Species)
Data: data
Control: glmerControl("bobyqa")
AIC BIC logLik -2*log(L) df.resid
1368.9 1423.4 -674.4 1348.9 1719
Scaled residuals:
Min 1Q Median 3Q Max
-2.5938 -0.2658 -0.1018 0.2001 3.3353
Random effects:
Groups Name Variance Std.Dev. Corr
Species (Intercept) 8.16321 2.8571
sAltitude 4.38183 2.0933 -0.13
Site (Intercept) 0.06786 0.2605
Number of obs: 1729, groups: Species, 152; Site, 17
Fixed effects:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -2.6284 0.4066 -6.465 1.02e-10 ***
sSeedMass -1.3693 0.6438 -2.127 0.0334 *
sSeedShape -0.5816 0.3100 -1.876 0.0606 .
sSeedN -2.1419 1.3184 -1.625 0.1042
sAltitude -1.3920 0.3362 -4.141 3.46e-05 ***
sHum -0.1022 0.1307 -0.781 0.4345
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Correlation of Fixed Effects:
(Intr) sSdMss sSdShp sSeedN sAlttd
sSeedMass 0.222
sSeedShape 0.033 0.062
sSeedN 0.337 0.061 -0.066
sAltitude 0.132 0.142 0.079 0.012
sHum 0.023 0.002 -0.003 -0.011 0.229
Success, it converged!
Residual checks:
Check residuals:
= simulateResiduals (model1, re.form= NULL , plot= TRUE )
Residuals look good!
Bonus: With phylogenetic correlation structure:
= ape:: cophenetic.phylo (phyl.upd2) # create distance matrix = vcv (phyl.upd2)[unique (data$ Species), unique (data$ Species)]### #the following code was taken from https://github.com/glmmTMB/glmmTMB/blob/master/misc/fixcorr.rmd <- function (Sigma,corrs.only= FALSE ) {<- log (diag (Sigma))/ 2 <- cov2cor (Sigma)<- chol (cr)<- cc %*% diag (1 / diag (cc))<- cc[upper.tri (cc)]if (corrs.only) return (corrs)<- c (logsd,corrs)return (ret)= as.theta.vcov (correlation_matrix, corrs.only= TRUE )##### $ dummy = factor (rep (0 , nrow (data)))= length (unique (data$ Species))= glmmTMB (SBPA~ + sSeedShape + sSeedN + + sHum + 1 | Site) + (sAltitude| Species) + 1 + Species| dummy),map= list (theta= factor (c (rep (0 , 4 ), rep (1 ,nsp),rep (NA ,length (corrs))) )),start= list (theta= c (rep (0 , 4 ), rep (0 ,nsp),corrs)),family = binomial,data = data)simulateResiduals (model6, plot = TRUE )
Object of Class DHARMa with simulated residuals based on 250 simulations with refit = FALSE . See ?DHARMa::simulateResiduals for help.
Scaled residual values: 0.5024676 0.5329717 0.5854716 0.9968902 0.7858493 0.5647619 0.1162222 0.1334808 0.1071773 0.3725763 0.2077203 0.3276229 0.02545432 0.06809947 0.3315729 0.3429219 0.6135937 0.4816801 0.6575116 0.8497154 ...
Conditional simulations:
= predict (model6, re.form = NULL , type = "response" )= sapply (1 : 1000 , function (i) rbinom (length (pred),1 , pred))= createDHARMa (simulations, model.frame (model6)[,1 ], pred)plot (res)
Snouter
Fit one of the responses in the snouter datset against the predictors rain + djungle (see ?snouter). Check for spatial autocorrelation and proceed to fitting a spatial model if needed. See the data set’s help for details on the variables.
library (EcoData)str (snouter)
Volcanoe Island
Option 2: Analyse ?volcanoisland in the EcoData package. Perform an appropriate causal statistical analysis (including residual checks and everything) to understand the predictors that determine the value of
windObs (numeric) lizards (1/0) beetles2 (counts) survived (k/n) There are various predictors, including plot, year, x, y that can be considered. Assume that all the issues that we talked about (RE, overdispersion, zero-inflation, spatial autocorrelation) could appear in this data.
Hint: it is possible that the possibility to observe a species depends on wind, and it is possible that lizards eat beetles, so you can also consider these variables (or predictions for these variables) as predictors of the others.
Hint 2: binomial k/n data in R is specified as a response that is cbind(k, n-k), so you would write glm(cbind(survived, all-survived) ~ predictor, data = data, family = “binomial”)